BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells
- PMID: 34282330
- PMCID: PMC8319109
- DOI: 10.1038/s41590-021-00964-8
BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells
Abstract
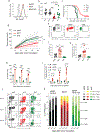
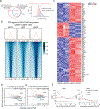
The transcription factors nuclear factor of activated T cells (NFAT) and activator protein 1 (AP-1; Fos-Jun) cooperate to promote the effector functions of T cells, but NFAT in the absence of AP-1 imposes a negative feedback program of T cell hyporesponsiveness (exhaustion). Here, we show that basic leucine zipper ATF-like transcription factor (BATF) and interferon regulatory factor 4 (IRF4) cooperate to counter T cell exhaustion in mouse tumor models. Overexpression of BATF in CD8+ T cells expressing a chimeric antigen receptor (CAR) promoted the survival and expansion of tumor-infiltrating CAR T cells, increased the production of effector cytokines, decreased the expression of inhibitory receptors and the exhaustion-associated transcription factor TOX and supported the generation of long-lived memory T cells that controlled tumor recurrence. These responses were dependent on BATF-IRF interaction, since cells expressing a BATF variant unable to interact with IRF4 did not survive in tumors and did not effectively delay tumor growth. BATF may improve the antitumor responses of CAR T cells by skewing their phenotypes and transcriptional profiles away from exhaustion and towards increased effector function.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures















Comment in
-
BATF targets T cell exhaustion for termination.Nat Immunol. 2021 Aug;22(8):936-938. doi: 10.1038/s41590-021-00978-2. Nat Immunol. 2021. PMID: 34282328 Free PMC article.
References
-
- McLane LM, Abdel-Hakeem MS & Wherry EJ CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 37, 457–495 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

