Deleterious Effects of SARS-CoV-2 Infection on Human Pancreatic Cells
- PMID: 34282405
- PMCID: PMC8285288
- DOI: 10.3389/fcimb.2021.678482
Deleterious Effects of SARS-CoV-2 Infection on Human Pancreatic Cells
Abstract
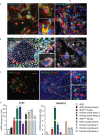
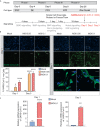
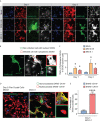
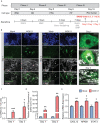
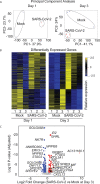
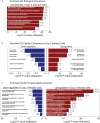
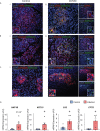
COVID-19 pandemic has infected more than 154 million people worldwide and caused more than 3.2 million deaths. It is transmitted by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and affects the respiratory tract as well as extra-pulmonary systems, including the pancreas, that express the virus entry receptor, Angiotensin-Converting Enzyme 2 (ACE2) receptor. Importantly, the endocrine and exocrine pancreas, the latter composed of ductal and acinar cells, express high levels of ACE2, which correlates to impaired functionality characterized as acute pancreatitis observed in some cases presenting with COVID-19. Since acute pancreatitis is already one of the most frequent gastrointestinal causes of hospitalization in the U.S. and the majority of studies investigating the effects of SARS-CoV-2 on the pancreas are clinical and observational, we utilized human iPSC technology to investigate the potential deleterious effects of SARS-CoV-2 infection on iPSC-derived pancreatic cultures containing endocrine and exocrine cells. Interestingly, iPSC-derived pancreatic cultures allow SARS-CoV-2 entry and establish infection, thus perturbing their normal molecular and cellular phenotypes. The infection increased a key cytokine, CXCL12, known to be involved in inflammatory responses in the pancreas. Transcriptome analysis of infected pancreatic cultures confirmed that SARS-CoV-2 hijacks the ribosomal machinery in these cells. Notably, the SARS-CoV-2 infectivity of the pancreas was confirmed in post-mortem tissues from COVID-19 patients, which showed co-localization of SARS-CoV-2 in pancreatic endocrine and exocrine cells and increased the expression of some pancreatic ductal stress response genes. Thus, we demonstrate that SARS-CoV-2 can directly infect human iPSC-derived pancreatic cells with strong supporting evidence of presence of the virus in post-mortem pancreatic tissue of confirmed COVID-19 human cases. This novel model of iPSC-derived pancreatic cultures will open new avenues for the comprehension of the SARS-CoV-2 infection and potentially establish a platform for endocrine and exocrine pancreas-specific antiviral drug screening.
Keywords: COVID-19; SARS-CoV-2; acinar cells; ductal cells; iPSCs; islets; pancreas; pancreatitis.
Copyright © 2021 Shaharuddin, Wang, Santos, Gross, Wang, Jawanda, Zhang, Hasan, Garcia, Arumugaswami and Sareen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
SARS-CoV-2 leading to acute pancreatitis: an unusual presentation.Braz J Infect Dis. 2020 Nov-Dec;24(6):561-564. doi: 10.1016/j.bjid.2020.08.011. Epub 2020 Sep 15. Braz J Infect Dis. 2020. PMID: 32961108 Free PMC article.
-
SARS-CoV-2 and the pancreas: What do we know about acute pancreatitis in COVID-19 positive patients?World J Gastroenterol. 2022 Sep 28;28(36):5240-5249. doi: 10.3748/wjg.v28.i36.5240. World J Gastroenterol. 2022. PMID: 36185634 Free PMC article.
-
Review on acute pancreatitis attributed to COVID-19 infection.World J Gastroenterol. 2022 May 21;28(19):2034-2056. doi: 10.3748/wjg.v28.i19.2034. World J Gastroenterol. 2022. PMID: 35664035 Free PMC article. Review.
-
SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas.Nat Metab. 2021 Feb;3(2):149-165. doi: 10.1038/s42255-021-00347-1. Epub 2021 Feb 3. Nat Metab. 2021. PMID: 33536639
-
Expression of SARS-CoV-2 receptor "ACE2" in human pancreatic β cells: to be or not to be!Islets. 2021 Sep 3;13(5-6):106-114. doi: 10.1080/19382014.2021.1954458. Epub 2021 Jul 24. Islets. 2021. PMID: 34304698 Free PMC article. Review.
Cited by
-
Digestive system infection by SARS‑CoV‑2: Entry mechanism, clinical symptoms and expression of major receptors (Review).Int J Mol Med. 2023 Mar;51(3):19. doi: 10.3892/ijmm.2023.5222. Epub 2023 Jan 20. Int J Mol Med. 2023. PMID: 36660939 Free PMC article. Review.
-
Association of COVID-19 with diabetes: a systematic review and meta-analysis.Sci Rep. 2022 Nov 23;12(1):20191. doi: 10.1038/s41598-022-24185-7. Sci Rep. 2022. PMID: 36418912 Free PMC article.
-
The Pathogenesis of Gastrointestinal, Hepatic, and Pancreatic Injury in Acute and Long Coronavirus Disease 2019 Infection.Gastroenterol Clin North Am. 2023 Mar;52(1):1-11. doi: 10.1016/j.gtc.2022.12.001. Epub 2022 Dec 5. Gastroenterol Clin North Am. 2023. PMID: 36813418 Free PMC article. Review.
-
New-Onset Diabetes Mellitus, Hypertension, Dyslipidaemia as Sequelae of COVID-19 Infection-Systematic Review.Int J Environ Res Public Health. 2022 Oct 14;19(20):13280. doi: 10.3390/ijerph192013280. Int J Environ Res Public Health. 2022. PMID: 36293857 Free PMC article.
-
The burden of acute pancreatitis on COVID-19 in the United States.Ann Gastroenterol. 2023 Mar-Apr;36(2):208-215. doi: 10.20524/aog.2023.0782. Epub 2023 Feb 3. Ann Gastroenterol. 2023. PMID: 36864935 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

