Kinetics of the Severe Acute Respiratory Syndrome Coronavirus 2 Antibody Response and Serological Estimation of Time Since Infection
- PMID: 34282461
- PMCID: PMC8420633
- DOI: 10.1093/infdis/jiab375
Kinetics of the Severe Acute Respiratory Syndrome Coronavirus 2 Antibody Response and Serological Estimation of Time Since Infection
Abstract
Background: Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces a complex antibody response that varies by orders of magnitude between individuals and over time.
Methods: We developed a multiplex serological test for measuring antibodies to 5 SARS-CoV-2 antigens and the spike proteins of seasonal coronaviruses. We measured antibody responses in cohorts of hospitalized patients and healthcare workers followed for up to 11 months after symptoms. A mathematical model of antibody kinetics was used to quantify the duration of antibody responses. Antibody response data were used to train algorithms for estimating time since infection.
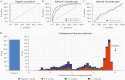
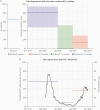
Results: One year after symptoms, we estimate that 36% (95% range, 11%-94%) of anti-Spike immunoglobulin G (IgG) remains, 31% (95% range, 9%-89%) anti-RBD IgG remains, and 7% (1%-31%) of anti-nucleocapsid IgG remains. The multiplex assay classified previous infections into time intervals of 0-3 months, 3-6 months, and 6-12 months. This method was validated using data from a seroprevalence survey in France, demonstrating that historical SARS-CoV-2 transmission can be reconstructed using samples from a single survey.
Conclusions: In addition to diagnosing previous SARS-CoV-2 infection, multiplex serological assays can estimate the time since infection, which can be used to reconstruct past epidemics.
Keywords: SARS-SoV-2; antibody kinetics; seroprevalence; surveillance; time since infection.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures


References
-
- Ward H, Cooke G, Atchison C, et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv [Preprint]. Posted online 27 October 2020. doi: 10.1101/2020.10.26.20219725. - DOI
-
- Kagucia EW, Gitonga JN, Kalu C, et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies among truck drivers and assistants in Kenya. medRxiv [Preprint]. Posted online 17 February 2021. doi: 10.1101/2021.02.12.21251294. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

