Machine Learning Methods for Fear Classification Based on Physiological Features
- PMID: 34282759
- PMCID: PMC8271969
- DOI: 10.3390/s21134519
Machine Learning Methods for Fear Classification Based on Physiological Features
Abstract
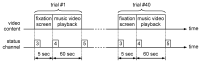
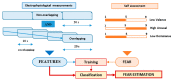
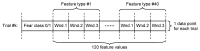
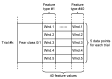
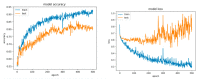
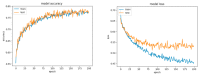
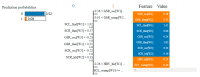
This paper focuses on the binary classification of the emotion of fear, based on the physiological data and subjective responses stored in the DEAP dataset. We performed a mapping between the discrete and dimensional emotional information considering the participants' ratings and extracted a substantial set of 40 types of features from the physiological data, which represented the input to various machine learning algorithms-Decision Trees, k-Nearest Neighbors, Support Vector Machine and artificial networks-accompanied by dimensionality reduction, feature selection and the tuning of the most relevant hyperparameters, boosting classification accuracy. The methodology we approached included tackling different situations, such as resolving the problem of having an imbalanced dataset through data augmentation, reducing overfitting, computing various metrics in order to obtain the most reliable classification scores and applying the Local Interpretable Model-Agnostic Explanations method for interpretation and for explaining predictions in a human-understandable manner. The results show that fear can be predicted very well (accuracies ranging from 91.7% using Gradient Boosting Trees to 93.5% using dimensionality reduction and Support Vector Machine) by extracting the most relevant features from the physiological data and by searching for the best parameters which maximize the machine learning algorithms' classification scores.
Keywords: emotion classification; emotion dimensions; fear classification; machine learning; neural networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

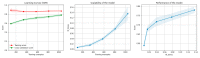


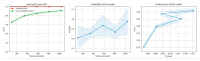



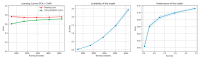
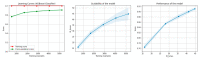

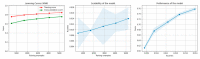

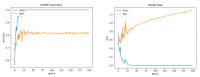
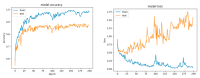

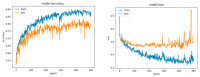




References
-
- Domínguez-Jiménez J., Campo-Landines K., Martínez-Santos J., Delahoz E., Contreras-Ortiz S. A machine learning model for emotion recognition from physiological signals. Biomed. Signal Process. Control. 2020;55:101646. doi: 10.1016/j.bspc.2019.101646. - DOI
-
- Ekman P. An argument for basic emotions. Cogn. Emot. 1992;6:169–200. doi: 10.1080/02699939208411068. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

