Comparative Genomics of Exiguobacterium Reveals What Makes a Cosmopolitan Bacterium
- PMID: 34282940
- PMCID: PMC8407118
- DOI: 10.1128/mSystems.00383-21
Comparative Genomics of Exiguobacterium Reveals What Makes a Cosmopolitan Bacterium
Abstract
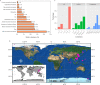
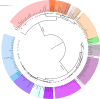
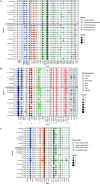
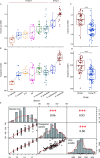
Although the strategies used by bacteria to adapt to specific environmental conditions are widely reported, fewer studies have addressed how microbes with a cosmopolitan distribution can survive in diverse ecosystems. Exiguobacterium is a versatile genus whose members are commonly found in various habitats. To better understand the mechanisms underlying the universality of Exiguobacterium, we collected 105 strains from diverse environments and performed large-scale metabolic and adaptive ability tests. We found that most Exiguobacterium members have the capacity to survive under wide ranges of temperature, salinity, and pH. According to phylogenetic and average nucleotide identity analyses, we identified 27 putative species and classified two genetic groups: groups I and II. Comparative genomic analysis revealed that the Exiguobacterium members utilize a variety of complex polysaccharides and proteins to support survival in diverse environments and also employ a number of chaperonins and transporters for this purpose. We observed that the group I species can be found in more diverse terrestrial environments and have a larger genome size than the group II species. Our analyses revealed that the expansion of transporter families drove genomic expansion in group I strains, and we identified 25 transporter families, many of which are involved in the transport of important substrates and resistance to environmental stresses and are enriched in group I strains. This study provides important insights into both the overall general genetic basis for the cosmopolitan distribution of a bacterial genus and the evolutionary and adaptive strategies of Exiguobacterium. IMPORTANCE The wide distribution characteristics of Exiguobacterium make it a valuable model for studying the adaptive strategies of bacteria that can survive in multiple habitats. In this study, we reveal that members of the Exiguobacterium genus have a cosmopolitan distribution and share an extensive adaptability that enables them to survive in various environments. The capacities shared by Exiguobacterium members, such as their diverse means of polysaccharide utilization and environmental-stress resistance, provide an important basis for their cosmopolitan distribution. Furthermore, the selective expansion of transporter families has been a main driving force for genomic evolution in Exiguobacterium. Our findings improve our understanding of the adaptive and evolutionary mechanisms of cosmopolitan bacteria and the vital genomic traits that can facilitate niche adaptation.
Keywords: Exiguobacterium; adaptation strategies; cosmopolitan distribution; genomics; polysaccharide utilization; transporters.
Figures

References
-
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Øvreås L, Reysenbach A-L, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi:10.1038/nrmicro1341. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

