Roles of homologous recombination in response to ionizing radiation-induced DNA damage
- PMID: 34283012
- PMCID: PMC9629169
- DOI: 10.1080/09553002.2021.1956001
Roles of homologous recombination in response to ionizing radiation-induced DNA damage
Abstract
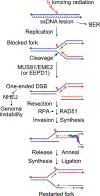
Purpose: Ionizing radiation induces a vast array of DNA lesions including base damage, and single- and double-strand breaks (SSB, DSB). DSBs are among the most cytotoxic lesions, and mis-repair causes small- and large-scale genome alterations that can contribute to carcinogenesis. Indeed, ionizing radiation is a 'complete' carcinogen. DSBs arise immediately after irradiation, termed 'frank DSBs,' as well as several hours later in a replication-dependent manner, termed 'secondary' or 'replication-dependent DSBs. DSBs resulting from replication fork collapse are single-ended and thus pose a distinct problem from two-ended, frank DSBs. DSBs are repaired by error-prone nonhomologous end-joining (NHEJ), or generally error-free homologous recombination (HR), each with sub-pathways. Clarifying how these pathways operate in normal and tumor cells is critical to increasing tumor control and minimizing side effects during radiotherapy.
Conclusions: The choice between NHEJ and HR is regulated during the cell cycle and by other factors. DSB repair pathways are major contributors to cell survival after ionizing radiation, including tumor-resistance to radiotherapy. Several nucleases are important for HR-mediated repair of replication-dependent DSBs and thus replication fork restart. These include three structure-specific nucleases, the 3' MUS81 nuclease, and two 5' nucleases, EEPD1 and Metnase, as well as three end-resection nucleases, MRE11, EXO1, and DNA2. The three structure-specific nucleases evolved at very different times, suggesting incremental acceleration of replication fork restart to limit toxic HR intermediates and genome instability as genomes increased in size during evolution, including the gain of large numbers of HR-prone repetitive elements. Ionizing radiation also induces delayed effects, observed days to weeks after exposure, including delayed cell death and delayed HR. In this review we highlight the roles of HR in cellular responses to ionizing radiation, and discuss the importance of HR as an exploitable target for cancer radiotherapy.
Keywords: DNA double-strand breaks; DNA repair; cancer radiotherapy; homologous recombination; ionizing radiation; replication stress.
Conflict of interest statement
Disclosure of interest
The authors report no conflict of interest.
Figures




References
-
- Allen C, Halbrook J, Nickoloff JA. 2003. Interactive competition between homologous recombination and non-homologous end joining. Mol Cancer Res. 1:913–920. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
