Obesity and diabetes are major risk factors for epicardial adipose tissue inflammation
- PMID: 34283808
- PMCID: PMC8409986
- DOI: 10.1172/jci.insight.145495
Obesity and diabetes are major risk factors for epicardial adipose tissue inflammation
Abstract
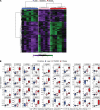
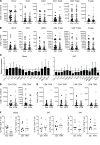
BACKGROUNDEpicardial adipose tissue (EAT) directly overlies the myocardium, with changes in its morphology and volume associated with myriad cardiovascular and metabolic diseases. However, EAT's immune structure and cellular characterization remain incompletely described. We aimed to define the immune phenotype of EAT in humans and compare such profiles across lean, obese, and diabetic patients.METHODSWe recruited 152 patients undergoing open-chest coronary artery bypass grafting (CABG), valve repair/replacement (VR) surgery, or combined CABG/VR. Patients' clinical and biochemical data and EAT, subcutaneous adipose tissue (SAT), and preoperative blood samples were collected. Immune cell profiling was evaluated by flow cytometry and complemented by gene expression studies of immune mediators. Bulk RNA-Seq was performed in EAT across metabolic profiles to assess whole-transcriptome changes observed in lean, obese, and diabetic groups.RESULTSFlow cytometry analysis demonstrated EAT was highly enriched in adaptive immune (T and B) cells. Although overweight/obese and diabetic patients had similar EAT cellular profiles to lean control patients, the EAT exhibited significantly (P ≤ 0.01) raised expression of immune mediators, including IL-1, IL-6, TNF-α, and IFN-γ. These changes were not observed in SAT or blood. Neither underlying coronary artery disease nor the presence of hypertension significantly altered the immune profiles observed. Bulk RNA-Seq demonstrated significant alterations in metabolic and inflammatory pathways in the EAT of overweight/obese patients compared with lean controls.CONCLUSIONAdaptive immune cells are the predominant immune cell constituent in human EAT and SAT. The presence of underlying cardiometabolic conditions, specifically obesity and diabetes, rather than cardiac disease phenotype appears to alter the inflammatory profile of EAT. Obese states markedly alter EAT metabolic and inflammatory signaling genes, underlining the impact of obesity on the EAT transcriptome profile.FUNDINGBarts Charity MGU0413, Abbott, Medical Research Council MR/T008059/1, and British Heart Foundation FS/13/49/30421 and PG/16/79/32419.
Keywords: Cardiology; Cardiovascular disease; Inflammation; Obesity; T cells.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

