CD8+PD-L1+CXCR3+ polyfunctional T cell abundances are associated with survival in critical SARS-CoV-2-infected patients
- PMID: 34283810
- PMCID: PMC8492305
- DOI: 10.1172/jci.insight.151571
CD8+PD-L1+CXCR3+ polyfunctional T cell abundances are associated with survival in critical SARS-CoV-2-infected patients
Abstract
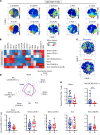
The importance of the adaptive T cell response in the control and resolution of viral infection has been well established. However, the nature of T cell-mediated viral control mechanisms in life-threatening stages of COVID-19 has yet to be determined. The aim of the present study was to determine the function and phenotype of T cell populations associated with survival or death of patients with COVID-19 in intensive care as a result of phenotypic and functional profiling by mass cytometry. Increased frequencies of circulating, polyfunctional CD4+CXCR5+HLA-DR+ stem cell memory T cells (Tscms) and decreased proportions of granzyme B-expressing and perforin-expressing effector memory T cells were detected in recovered and deceased patients, respectively. The higher abundance of polyfunctional PD-L1+CXCR3+CD8+ effector T cells (Teffs), CXCR5+HLA-DR+ Tscms, and anti-nucleocapsid (anti-NC) cytokine-producing T cells permitted us to differentiate between recovered and deceased patients. The results from a principal component analysis show an imbalance in the T cell compartment that allowed for the separation of recovered and deceased patients. The paucity of circulating PD-L1+CXCR3+CD8+ Teffs and NC-specific CD8+ T cells accurately forecasts fatal disease outcome. This study provides insight into the nature of the T cell populations involved in the control of COVID-19 and therefore might impact T cell-based vaccine designs for this infectious disease.
Keywords: Adaptive immunity; Immunology; T cells.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

