Clinical significance and mechanisms associated with segmental UPD
- PMID: 34284807
- PMCID: PMC8290618
- DOI: 10.1186/s13039-021-00555-0
Clinical significance and mechanisms associated with segmental UPD
Abstract
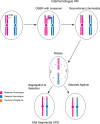
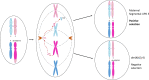
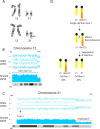
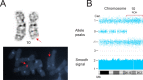
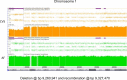
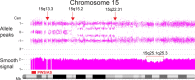
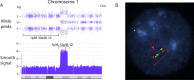
Whole chromosome uniparental disomy (UPD) has been well documented with mechanisms largely understood. However, the etiology of segmental limited UPD (segUPD) is not as clear. In a 10-year period of confirming (> 300) cases of whole chromosome UPD, we identified 86 segmental cases in both prenatal and postnatal samples. Thirty-two of these cases showed mosaic segmental UPD at 11p due to somatic selection associated with Beckwith-Wiedemann syndrome. This study focuses on apparent mechanisms associated with the remaining cases, many of which appear to represent corrections of genomic imbalance such as deletions and derivative chromosomes. In some cases, segmental UPD was associated with the generation of additional genomic imbalance while in others it apparently resulted in restoration of euploidy. Multiple tests utilizing noninvasive prenatal testing (NIPT), chorionic villus sampling (CVS) and amniotic fluid samples from the same pregnancy revealed temporal evidence of correction and a "hotspot" at 1p. Although in many cases the genomic imbalance was dosage "repaired" in the analyzed tissue, clinical effects could be sustained due to early developmental effects of the original imbalance or due to its continued existence in other tissues. In addition, if correction did not occur in the gametes there would be recurrence risks for the offspring of those individuals. Familial microarray allele patterns are presented that differentiate lack of gamete correction from somatic derived gonadal mosaicism. These results suggest that the incidence of segUPD mediated correction is underestimated and may explain the etiology of some clinical phenotypes which are undetected by routine microarray analysis and many exome sequencing studies.
Keywords: Chromosome microarray (CMA); Cytogenetics; Homologous recombination; Mitotic correction; Segmental uniparental disomy; Uniparental disomy.
© 2021. The Author(s).
Conflict of interest statement
Authors PP, CK, SC, SS, and AP are employees of Laboratory Corporation of America with option to hold stock. SH is employed by University of South Florida.
Figures







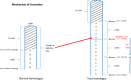
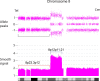
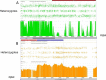

Similar articles
-
Complex and segmental uniparental disomy (UPD): review and lessons from rare chromosomal complements.J Med Genet. 2001 Aug;38(8):497-507. doi: 10.1136/jmg.38.8.497. J Med Genet. 2001. PMID: 11483637 Free PMC article. Review.
-
Mitotic recombination and uniparental disomy in Beckwith-Wiedemann syndrome.Genomics. 2007 May;89(5):613-7. doi: 10.1016/j.ygeno.2007.01.005. Epub 2007 Mar 6. Genomics. 2007. PMID: 17337339
-
Identification of consensus motifs associated with mitotic recombination and clinical characteristics in patients with paternal uniparental isodisomy of chromosome 11.Hum Mol Genet. 2016 Apr 1;25(7):1406-19. doi: 10.1093/hmg/ddw023. Epub 2016 Jan 28. Hum Mol Genet. 2016. PMID: 26908620
-
Complex and segmental uniparental disomy updated.J Med Genet. 2008 Sep;45(9):545-56. doi: 10.1136/jmg.2008.058016. Epub 2008 Jun 4. J Med Genet. 2008. PMID: 18524837 Review.
-
Prenatal testing for uniparental disomy: indications and clinical relevance.Ultrasound Obstet Gynecol. 2008 Jan;31(1):100-5. doi: 10.1002/uog.5133. Ultrasound Obstet Gynecol. 2008. PMID: 18059071 Review.
Cited by
-
Uniparental IsoDisomy: a case study on a new mechanism of Friedreich ataxia.Eur J Hum Genet. 2025 Jan;33(1):137-140. doi: 10.1038/s41431-024-01728-2. Epub 2024 Nov 4. Eur J Hum Genet. 2025. PMID: 39496895
-
Case Report: Decrypting an interchromosomal insertion associated with Marfan's syndrome: how optical genome mapping emphasizes the morbid burden of copy-neutral variants.Front Genet. 2023 Sep 21;14:1244983. doi: 10.3389/fgene.2023.1244983. eCollection 2023. Front Genet. 2023. PMID: 37811140 Free PMC article.
-
Biliverdinuria Caused by Exonic BLVRA Deletions in Two Dogs with Green Urine.Genes (Basel). 2024 Nov 30;15(12):1561. doi: 10.3390/genes15121561. Genes (Basel). 2024. PMID: 39766828 Free PMC article.
-
An Incidental Detection of a Rare UPD in SNP-Array Based PGT-SR: A Case Report.Reprod Sci. 2024 Sep;31(9):2893-2899. doi: 10.1007/s43032-024-01598-5. Epub 2024 May 23. Reprod Sci. 2024. PMID: 38780745
References
LinkOut - more resources
Full Text Sources

