Engineering self-organized criticality in living cells
- PMID: 34285228
- PMCID: PMC8292319
- DOI: 10.1038/s41467-021-24695-4
Engineering self-organized criticality in living cells
Erratum in
-
Author Correction: Engineering self-organized criticality in living cells.Nat Commun. 2021 Sep 23;12(1):5699. doi: 10.1038/s41467-021-25603-6. Nat Commun. 2021. PMID: 34556646 Free PMC article. No abstract available.
Abstract
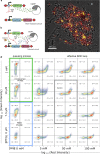
Complex dynamical fluctuations, from intracellular noise, brain dynamics or computer traffic display bursting dynamics consistent with a critical state between order and disorder. Living close to the critical point has adaptive advantages and it has been conjectured that evolution could select these critical states. Is this the case of living cells? A system can poise itself close to the critical point by means of the so-called self-organized criticality (SOC). In this paper we present an engineered gene network displaying SOC behaviour. This is achieved by exploiting the saturation of the proteolytic degradation machinery in E. coli cells by means of a negative feedback loop that reduces congestion. Our critical motif is built from a two-gene circuit, where SOC can be successfully implemented. The potential implications for both cellular dynamics and behaviour are discussed.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

