Integration strategies of multi-omics data for machine learning analysis
- PMID: 34285775
- PMCID: PMC8258788
- DOI: 10.1016/j.csbj.2021.06.030
Integration strategies of multi-omics data for machine learning analysis
Abstract
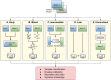
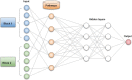
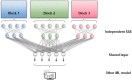
Increased availability of high-throughput technologies has generated an ever-growing number of omics data that seek to portray many different but complementary biological layers including genomics, epigenomics, transcriptomics, proteomics, and metabolomics. New insight from these data have been obtained by machine learning algorithms that have produced diagnostic and classification biomarkers. Most biomarkers obtained to date however only include one omic measurement at a time and thus do not take full advantage of recent multi-omics experiments that now capture the entire complexity of biological systems. Multi-omics data integration strategies are needed to combine the complementary knowledge brought by each omics layer. We have summarized the most recent data integration methods/ frameworks into five different integration strategies: early, mixed, intermediate, late and hierarchical. In this mini-review, we focus on challenges and existing multi-omics integration strategies by paying special attention to machine learning applications.
Keywords: Deep learning; Integration strategy; Machine learning; Multi-omics; Multi-view; Network.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Algorithms and tools for data-driven omics integration to achieve multilayer biological insights: a narrative review.J Transl Med. 2025 Apr 10;23(1):425. doi: 10.1186/s12967-025-06446-x. J Transl Med. 2025. PMID: 40211300 Free PMC article. Review.
-
A Customizable Analysis Flow in Integrative Multi-Omics.Biomolecules. 2020 Nov 27;10(12):1606. doi: 10.3390/biom10121606. Biomolecules. 2020. PMID: 33260881 Free PMC article. Review.
-
Statistical and Machine-Learning Analyses in Nutritional Genomics Studies.Nutrients. 2020 Oct 14;12(10):3140. doi: 10.3390/nu12103140. Nutrients. 2020. PMID: 33066636 Free PMC article. Review.
-
Integrating Molecular Perspectives: Strategies for Comprehensive Multi-Omics Integrative Data Analysis and Machine Learning Applications in Transcriptomics, Proteomics, and Metabolomics.Biology (Basel). 2024 Oct 22;13(11):848. doi: 10.3390/biology13110848. Biology (Basel). 2024. PMID: 39596803 Free PMC article. Review.
-
Multi-modal intermediate integrative methods in neuropsychiatric disorders: A review.Comput Struct Biotechnol J. 2022 Nov 8;20:6149-6162. doi: 10.1016/j.csbj.2022.11.008. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36420153 Free PMC article. Review.
Cited by
-
Transforming Clinical Research: The Power of High-Throughput Omics Integration.Proteomes. 2024 Sep 6;12(3):25. doi: 10.3390/proteomes12030025. Proteomes. 2024. PMID: 39311198 Free PMC article. Review.
-
HiOmics: A cloud-based one-stop platform for the comprehensive analysis of large-scale omics data.Comput Struct Biotechnol J. 2024 Jan 5;23:659-668. doi: 10.1016/j.csbj.2024.01.002. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38292471 Free PMC article.
-
Computational Methods for Single-Cell Imaging and Omics Data Integration.Front Mol Biosci. 2022 Jan 17;8:768106. doi: 10.3389/fmolb.2021.768106. eCollection 2021. Front Mol Biosci. 2022. PMID: 35111809 Free PMC article. Review.
-
Interpretable meta-learning of multi-omics data for survival analysis and pathway enrichment.Bioinformatics. 2023 Apr 3;39(4):btad113. doi: 10.1093/bioinformatics/btad113. Bioinformatics. 2023. PMID: 36864611 Free PMC article.
-
A guide to multi-omics data collection and integration for translational medicine.Comput Struct Biotechnol J. 2022 Dec 1;21:134-149. doi: 10.1016/j.csbj.2022.11.050. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36544480 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
