Heterologous Expression and Purification of CRISPR-Cas12a/Cpf1
- PMID: 34286046
- PMCID: PMC8275269
- DOI: 10.21769/BioProtoc.2842
Heterologous Expression and Purification of CRISPR-Cas12a/Cpf1
Abstract
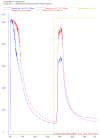
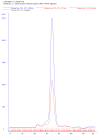
This protocol provides step by step instructions (Figure 1) for heterologous expression of Francisella novicida Cas12a (previously known as Cpf1) in Escherichia coli. It additionally includes a protocol for high-purity purification and briefly describes how activity assays can be performed. These protocols can also be used for purification of other Cas12a homologs and the purified proteins can be used for subsequent genome editing experiments. Figure 1. Timeline of activities for the heterologous expression and purification of Francisella novicida Cas12a (FnCas12a) from Escherichia coli.
Keywords: CRISPR-Cas; Cas12a; Cpf1; Protein purification.
Copyright © 2018 The Authors; exclusive licensee Bio-protocol LLC.
Figures







References
-
- Doudna J. A. and Charpentier E.(2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213): 1258096. - PubMed
LinkOut - more resources
Full Text Sources

