Single-cell image analysis reveals a protective role for microglia in glioblastoma
- PMID: 34286275
- PMCID: PMC8284623
- DOI: 10.1093/noajnl/vdab031
Single-cell image analysis reveals a protective role for microglia in glioblastoma
Abstract
Background: Microglia and tumor-associated macrophages (TAMs) constitute up to half of the total tumor mass of glioblastomas. Despite these myeloid populations being ontogenetically distinct, they have been largely conflated. Recent single-cell transcriptomic studies have identified genes that distinguish microglia from TAMs. Here we investigated whether the translated proteins of genes enriched in microglial or TAM populations can be used to differentiate these myeloid cells in immunohistochemically stained human glioblastoma tissue.
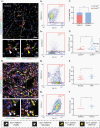
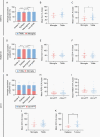
Methods: Tissue sections from resected low-grade, meningioma, and glioblastoma (grade IV) tumors and epilepsy tissues were immunofluorescently triple-labeled for Iba1 (pan-myeloid marker), CD14 or CD163 (preferential TAM markers), and either P2RY12 or TMEM119 (microglial-specific markers). Using a single-cell-based image analysis pipeline, we quantified the abundance of each marker within single myeloid cells, allowing the identification and analysis of myeloid populations.
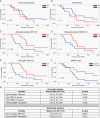
Results: P2RY12 and TMEM119 successfully discriminated microglia from TAMs in glioblastoma. In contrast, CD14 and CD163 expression were not restricted to invading TAMs and were upregulated by tumor microglia. Notably, a higher ratio of microglia to TAMs significantly correlated with increased patient survival.
Conclusions: We demonstrate the validity of previously defined microglial-specific genes P2RY12 and TMEM119 as robust discriminators of microglia and TAMs at the protein level in human tissue. Moreover, our data suggest that a higher proportion of microglia may be beneficial for patient survival in glioblastoma. Accordingly, this tissue-based method for myeloid population differentiation could serve as a useful prognostic tool.
Keywords: immunosuppression; microglia; tumor immunology; tumor-associated macrophages.
© The Author(s) 2021. Published by Oxford University Press, the Society for Neuro-Oncology and the European Association of Neuro-Oncology.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
