Establishment of a Patient-Derived, Magnetic Levitation-Based, Three-Dimensional Spheroid Granuloma Model for Human Tuberculosis
- PMID: 34287004
- PMCID: PMC8386456
- DOI: 10.1128/mSphere.00552-21
Establishment of a Patient-Derived, Magnetic Levitation-Based, Three-Dimensional Spheroid Granuloma Model for Human Tuberculosis
Abstract
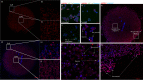
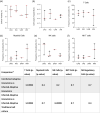
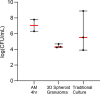
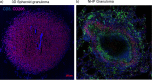
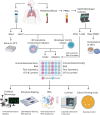
Tuberculous granulomas that develop in response to Mycobacterium tuberculosis (M. tuberculosis) infection are highly dynamic entities shaped by the host immune response and disease kinetics. Within this microenvironment, immune cell recruitment, polarization, and activation are driven not only by coexisting cell types and multicellular interactions but also by M. tuberculosis-mediated changes involving metabolic heterogeneity, epigenetic reprogramming, and rewiring of the transcriptional landscape of host cells. There is an increased appreciation of the in vivo complexity, versatility, and heterogeneity of the cellular compartment that constitutes the tuberculosis (TB) granuloma and the difficulty in translating findings from animal models to human disease. Here, we describe a novel biomimetic in vitro three-dimensional (3D) human lung spheroid granuloma model, resembling early "innate" and "adaptive" stages of the TB granuloma spectrum, and present results of histological architecture, host transcriptional characterization, mycobacteriological features, cytokine profiles, and spatial distribution of key immune cells. A range of manipulations of immune cell populations in these spheroid granulomas will allow the study of host/pathogen pathways involved in the outcome of infection, as well as pharmacological interventions. IMPORTANCE TB is a highly infectious disease, with granulomas as its hallmark. Granulomas play an important role in the control of M. tuberculosis infection and as such are crucial indicators for our understanding of host resistance to TB. Correlates of risk and protection to M. tuberculosis are still elusive, and the granuloma provides the perfect environment in which to study the immune response to infection and broaden our understanding thereof; however, human granulomas are difficult to obtain, and animal models are costly and do not always faithfully mimic human immunity. In fact, most TB research is conducted in vitro on immortalized or primary immune cells and cultured in two dimensions on flat, rigid plastic, which does not reflect in vivo characteristics. We have therefore conceived a 3D, human in vitro spheroid granuloma model which allows researchers to study features of granuloma-forming diseases in a 3D structural environment resembling in vivo granuloma architecture and cellular orientation.
Keywords: 3D cell culture; granuloma; spheroid; tuberculosis.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

