Intragenic Distribution of IS 6110 in Clinical Mycobacterium tuberculosis Strains: Bioinformatic Evidence for Gene Disruption Leading to Underdiagnosed Antibiotic Resistance
- PMID: 34287057
- PMCID: PMC8552512
- DOI: 10.1128/Spectrum.00019-21
Intragenic Distribution of IS 6110 in Clinical Mycobacterium tuberculosis Strains: Bioinformatic Evidence for Gene Disruption Leading to Underdiagnosed Antibiotic Resistance
Abstract
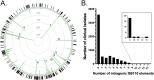
Antibiotic resistance is a global challenge for tuberculosis control, and accelerating its diagnosis is critical for therapy decisions and controlling transmission. Genotype-based molecular diagnostics now play an increasing role in accelerating the detection of such antibiotic resistance, but their accuracy depends on the instructed detection of genetic variations. Genetic mobile elements such as IS6110 are established sources of genetic variation in Mycobacterium tuberculosis, but their implication in clinical antibiotic resistance has thus far been unclear. Here, we describe the discovery of an intragenic IS6110 insertion into Rv0678 that caused antibiotic resistance in an in vitro-selected M. tuberculosis isolate. The subsequent development of bioinformatics scripts allowed genome-wide analysis of intragenic IS6110 insertions causing gene disruptions in 6,426 clinical M. tuberculosis strains. This analysis identified 10,070 intragenic IS6110 insertions distributed among 333 different genes. Focusing on genes whose disruption leads to antibiotic resistance, 12 clinical isolates were identified with high confidence to be resistant to bedaquiline, clofazimine, pyrazinamide, ethionamide, and para-aminosalicylic acid because of an IS6110-mediated gene disruption event. A number of these IS6110-mediated resistant strains had identical genomic distributions of IS6110 elements and likely represent transmission events of a single resistant isolate. These data provide strong evidence that IS6110-mediated gene disruption is a clinically relevant mechanism of antibiotic resistance in M. tuberculosis that should be considered for molecular diagnostics. Concomitantly, this analysis provides a list of 333 IS6110-disrupted genes in clinical tuberculosis isolates that can be deemed nonessential for human infection. IMPORTANCE To help control the spread of drug-resistant tuberculosis and to guide treatment choices, it is important that rapid and accurate molecular diagnostic tools are used. Current molecular diagnostic tools detect the most common antibiotic-resistance-conferring mutations in the form of single nucleotide changes, small deletions, or insertions. Mobile genetic elements, named IS6110, are also known to move within the M. tuberculosis genome and cause significant genetic variations, although the role of this variation in clinical drug resistance remains unclear. In this work, we show that both in vitro and in data analyzed from 6,426 clinical M. tuberculosis strains, IS6110 elements are found that disrupt specific genes essential for the function of a number of pivotal antituberculosis drugs. By providing ample evidence of clinically relevant IS6110-mediated drug resistance, we believe that this shows that this form of genetic variation must not be overlooked in molecular diagnostics of drug resistance.
Keywords: IS6110; Mycobacterium tuberculosis; antibiotic resistance; diagnostics; genetic polymorphisms.
Figures

References
-
- World Health Organization. 2020. Global tuberculosis report 2020. World Health Organization, Geneva, Switzerland.
-
- Bricogne G, Blanc E, Brandl M, Flensburg C, Keller PP, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. 2017. BUSTER version 2.10.3. Global Phasing Ltd, Cambridge, United Kingdom.
-
- DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi:10.1128/mBio.02133-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

