Genomic Update of Phenotypic Prediction Rule for Methicillin-Resistant Staphylococcus aureus (MRSA) USA300 Discloses Jail Transmission Networks with Increased Resistance
- PMID: 34287060
- PMCID: PMC8552710
- DOI: 10.1128/Spectrum.00376-21
Genomic Update of Phenotypic Prediction Rule for Methicillin-Resistant Staphylococcus aureus (MRSA) USA300 Discloses Jail Transmission Networks with Increased Resistance
Abstract
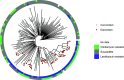
Methicillin-resistant Staphylococcus aureus (MRSA) is a leading cause of health care-associated (HA) and community-associated (CA) infections. USA300 strains are historically CA-MRSA, while USA100 strains are HA-MRSA. Here, we update an antibiotic prediction rule to distinguish these two genotypes based on antibiotic resistance phenotype using whole-genome sequencing (WGS), a more discriminatory methodology than pulsed-field gel electrophoresis (PFGE). MRSA clinical isolates collected from 2007 to 2017 underwent WGS; associated epidemiologic data were ascertained. In developing the rule, we examined MRSA isolates that included a population with a history of incarceration. Performance characteristics of antibiotic susceptibility for predicting USA300 compared to USA100, as defined by WGS, were examined. Phylogenetic analysis was performed to examine resistant USA300 clades. We identified 275 isolates (221 USA300, 54 USA100). Combination susceptibility to clindamycin or levofloxacin performed the best overall (sensitivity 80.7%, specificity 75.9%) to identify USA300. The average number of antibiotic classes with resistance was higher for USA100 (3 versus 2, P < 0.001). Resistance to ≤2 classes was predictive for USA300 (area under the curve (AUC) 0.84, 95% confidence interval 0.78 to 0.90). Phylogenetic analysis identified a cluster of USA300 strains characterized by increased resistance among incarcerated individuals. Using a combination of clindamycin or levofloxacin susceptibility, or resistance to ≤2 antibiotic classes, was predictive of USA300 as defined by WGS. Increased resistance was observed among individuals with incarceration exposure, suggesting circulation of a more resistant USA300 clade among at-risk community networks. Our phenotypic prediction rule could be used as an epidemiologic tool to describe community and nosocomial shifts in USA300 MRSA and quickly identify emergence of lineages with increased resistance. IMPORTANCE Methicillin-resistant Staphylococcus aureus (MRSA) is an important cause of health care-associated (HA) and community-associated (CA) infections, but the epidemiology of these strains (USA100 and USA300, respectively) now overlaps in health care settings. Although sequencing technology has become more available, many health care facilities still lack the capabilities to perform these analyses. In this study, we update a simple prediction rule based on antibiotic resistance phenotype with integration of whole-genome sequencing (WGS) to predict strain type based on antibiotic resistance profiles that can be used in settings without access to molecular strain typing methods. This prediction rule has many potential epidemiologic applications, such as analysis of retrospective data sets, regional monitoring, and ongoing surveillance of CA-MRSA infection trends. We demonstrate application of this rule to identify an emerging USA300 strain with increased antibiotic resistance among incarcerated individuals that deviates from the rule.
Keywords: MRSA; Staphylococcus aureus; antibiotic resistance; methicillin resistance.
Figures

Similar articles
-
Demography and Intercontinental Spread of the USA300 Community-Acquired Methicillin-Resistant Staphylococcus aureus Lineage.mBio. 2016 Feb 16;7(1):e02183-15. doi: 10.1128/mBio.02183-15. mBio. 2016. PMID: 26884428 Free PMC article.
-
USA300 methicillin-resistant Staphylococcus aureus in Stockholm, Sweden, from 2008 to 2016.PLoS One. 2018 Nov 7;13(11):e0205761. doi: 10.1371/journal.pone.0205761. eCollection 2018. PLoS One. 2018. PMID: 30403684 Free PMC article.
-
Pediatric Staphylococcus aureus Isolate Genotypes and Infections from the Dawn of the Community-Associated Methicillin-Resistant S. aureus Epidemic Era in Chicago, 1994 to 1997.J Clin Microbiol. 2015 Aug;53(8):2486-91. doi: 10.1128/JCM.00096-15. Epub 2015 May 27. J Clin Microbiol. 2015. PMID: 26019202 Free PMC article.
-
The evolution of Staphylococcus aureus.Infect Genet Evol. 2008 Dec;8(6):747-63. doi: 10.1016/j.meegid.2008.07.007. Epub 2008 Jul 29. Infect Genet Evol. 2008. PMID: 18718557 Review.
-
Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA).Curr Opin Microbiol. 2012 Oct;15(5):588-95. doi: 10.1016/j.mib.2012.08.003. Epub 2012 Oct 5. Curr Opin Microbiol. 2012. PMID: 23044073 Review.
Cited by
-
Progress in the Prevalence, Classification and Drug Resistance Mechanisms of Methicillin-Resistant Staphylococcus aureus.Infect Drug Resist. 2023 May 25;16:3271-3292. doi: 10.2147/IDR.S412308. eCollection 2023. Infect Drug Resist. 2023. PMID: 37255882 Free PMC article. Review.
-
Clonal Diversity, Antibiotic Resistance, and Virulence Factor Prevalence of Community Associated Staphylococcus aureus in Southeastern Virginia.Pathogens. 2023 Dec 27;13(1):25. doi: 10.3390/pathogens13010025. Pathogens. 2023. PMID: 38251333 Free PMC article.
-
Correlation Analysis of Staphylococcus aureus Drug Resistance and Virulence Factors with Blood Cell Counts and Coagulation Indexes.Int J Clin Pract. 2023 Feb 15;2023:8768152. doi: 10.1155/2023/8768152. eCollection 2023. Int J Clin Pract. 2023. PMID: 36846497 Free PMC article.
-
Staphylococcus aureus Fibronectin-Binding Proteins Contribute to Colonization of the Female Reproductive Tract.Infect Immun. 2023 Jan 24;91(1):e0046022. doi: 10.1128/iai.00460-22. Epub 2022 Dec 13. Infect Immun. 2023. PMID: 36511703 Free PMC article.
-
The mutational landscape of Staphylococcus aureus during colonisation.Nat Commun. 2025 Jan 13;16(1):302. doi: 10.1038/s41467-024-55186-x. Nat Commun. 2025. PMID: 39805814 Free PMC article.
References
-
- Roberts RB, Chung M, de Lencastre H, Hargrave J, Tomasz A, Nicolau DP, John JF, Jr, Korzeniowski O, Tri-State MRSA Collaborative Study Group. 2000. Distribution of methicillin-resistant Staphylococcus aureus clones among health care facilities in Connecticut, New Jersey, and Pennsylvania. Microb Drug Resist 6:245–251. doi:10.1089/mdr.2000.6.245. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

