Abnormal methylation characteristics predict chemoresistance and poor prognosis in advanced high-grade serous ovarian cancer
- PMID: 34289901
- PMCID: PMC8296752
- DOI: 10.1186/s13148-021-01133-2
Abnormal methylation characteristics predict chemoresistance and poor prognosis in advanced high-grade serous ovarian cancer
Abstract
Background: Primary or acquired chemoresistance is a key link in the high mortality rate of ovarian cancer. There is no reliable method to predict chemoresistance in ovarian cancer. We hypothesized that specific methylation characteristics could distinguish chemoresistant and chemosensitive ovarian cancer patients.
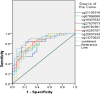
Methods: In this study, we used 450 K Infinium Methylation BeadChip to detect the different methylation CpGs between ovarian cancer patients. The differential methylation genes were analyzed by GO and KEGG Pathway bioinformatics analysis. The candidate CpGs were confirmed by pyrosequencing. The expression of abnormal methylation gene was identified by QRT-PCR and IHC. ROC analysis confirmed the ability to predict chemotherapy outcomes. Prognosis was evaluated using Kaplan-Meier.
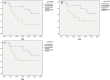
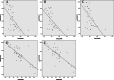
Results: In advanced high-grade serous ovarian cancer, 8 CpGs (ITGB6:cg21105318, cg07896068, cg18437633; NCALD: cg27637873, cg26782361, cg16265707; LAMA3: cg20937934, cg13270625) remained hypermethylated in chemoresistant patients. The sensitivity, specificity and AUC of 8 CpGs (ITGB6:cg21105318, cg07896068, cg18437633; NCALD: cg27637873, cg26782361, cg16265707; LAMA3: cg20937934, cg13270625) methylation to predict chemotherapy sensitivity were 63.60-97.00%, 46.40-89.30% and 0.774-0.846. PFS of 6 candidate genes (ITGB6:cg21105318, cg07896068; NCALD: cg27637873, cg26782361, cg16265707; LAMA3: cg20937934) hypermethylation patients was significantly shorter. The expression of NCALD and LAMA3 in chemoresistant patients was lower than that of chemosensitive patients. Spearman analysis showed that NCALD and LAMA3 methylations were negatively correlated with their expression.
Conclusions: As a new biomarker of chemotherapy sensitivity, hypermethylation of NCALD and LAMA3 is associated with poor PFS in advanced high-grade serous ovarian cancer. In the future, further research on NCALD and LAMA3 will be needed to provide guidance for clinical stratification of demethylation therapy.
Keywords: 450 K Infinium Methylation BeadChip; Chemotherapy resistance; DNA methylation; Ovarian cancer; Prognosis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
LAMA3 DNA methylation and transcriptome changes associated with chemotherapy resistance in ovarian cancer.J Ovarian Res. 2021 May 15;14(1):67. doi: 10.1186/s13048-021-00807-y. J Ovarian Res. 2021. PMID: 33992120 Free PMC article.
-
Low expression of NCALD is associated with chemotherapy resistance and poor prognosis in epithelial ovarian cancer.J Ovarian Res. 2020 Mar 30;13(1):35. doi: 10.1186/s13048-020-00635-6. J Ovarian Res. 2020. PMID: 32228639 Free PMC article.
-
[Clinical and bioinformatics analysis of the relationship between LAMA3 DNA methylation expression and platinum resistance and prognosis in epithelial ovarian cancer].Zhonghua Fu Chan Ke Za Zhi. 2024 Jun 25;59(6):454-464. doi: 10.3760/cma.j.cn112141-20240201-00069. Zhonghua Fu Chan Ke Za Zhi. 2024. PMID: 38951081 Chinese.
-
Hypermethylation of tumor suppressor genes is a risk factor for poor prognosis in ovarian cancer: A meta-analysis.Medicine (Baltimore). 2019 Feb;98(8):e14588. doi: 10.1097/MD.0000000000014588. Medicine (Baltimore). 2019. PMID: 30813180 Free PMC article. Review.
-
DNA Methylation in Epithelial Ovarian Cancer: Current Data and Future Perspectives.Curr Mol Pharmacol. 2021;14(6):1013-1027. doi: 10.2174/1874467213666200810141858. Curr Mol Pharmacol. 2021. PMID: 32778046
Cited by
-
Deciphering the Molecular Mechanisms behind Drug Resistance in Ovarian Cancer to Unlock Efficient Treatment Options.Cells. 2024 May 4;13(9):786. doi: 10.3390/cells13090786. Cells. 2024. PMID: 38727322 Free PMC article. Review.
-
The Methylation Game: Epigenetic and Epitranscriptomic Dynamics of 5-Methylcytosine.Front Cell Dev Biol. 2022 Jun 3;10:915685. doi: 10.3389/fcell.2022.915685. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35721489 Free PMC article. Review.
-
Comprehensive Analysis of DNA Methylation and Transcriptome to Identify PD-1-Negative Prognostic Methylated Signature in Endometrial Carcinoma.Dis Markers. 2022 May 18;2022:3085289. doi: 10.1155/2022/3085289. eCollection 2022. Dis Markers. 2022. PMID: 35634444 Free PMC article.
-
PCDH17 gene promoter methylation status in a cohort of Egyptian women with epithelial ovarian cancer.BMC Cancer. 2023 Jan 25;23(1):89. doi: 10.1186/s12885-023-10549-3. BMC Cancer. 2023. PMID: 36698136 Free PMC article.
-
Computational Analysis of High-Dimensional DNA Methylation Data for Cancer Prognosis.J Comput Biol. 2022 Aug;29(8):769-781. doi: 10.1089/cmb.2022.0002. Epub 2022 Jun 6. J Comput Biol. 2022. PMID: 35671506 Free PMC article. Review.
References
-
- Li Y, Wang ZC, Luo L, et al. The clinical value of the combined detection of sEGFR, CA125 and HE4 for epithelial ovarian cancer diagnosis. Eur Rev Med Pharmacol Sci. 2020;24(2):604–610. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

