rpoS-mutation variants are selected in Pseudomonas aeruginosa biofilms under imipenem pressure
- PMID: 34289907
- PMCID: PMC8293535
- DOI: 10.1186/s13578-021-00655-9
rpoS-mutation variants are selected in Pseudomonas aeruginosa biofilms under imipenem pressure
Abstract
Background: Pseudomonas aeruginosa is a notorious opportunistic pathogen causing various types of biofilm-related infections. Biofilm formation is a unique microbial strategy that allows P. aeruginosa to survive adverse conditions such as antibiotic treatment and human immune clearance.
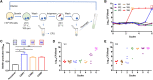
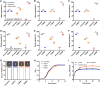
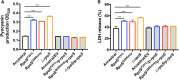
Results: In this study, we experimentally evolved P. aeruginosa PAO1 biofilms for cyclic treatment in the presence of high dose of imipenem, and enriched hyperbiofilm mutants within six cycles in two independent lineages. The competition assay showed that the evolved hyperbiofilm mutants can outcompete the ancestral strain within biofilms but not in planktonic cultures. Whole-genome sequencing analysis revealed the hyperbiofilm phenotype is caused by point mutations in rpoS gene in all independently evolved mutants and the same mutation was found in P. aeruginosa clinical isolates. We further showed that mutation in rpoS gene increased the intracellular c-di-GMP level by turning on the expression of the diguanylate cyclases. Mutation in rpoS increased pyocyanin production and virulence in hyperbiofilm variants.
Conclusion: Here, our study revealed that antibiotic treatment of biofilm-related P. aeruginosa infections might induce a hyperbiofilm phenotype via rpoS mutation, which might partially explain antimicrobial treatment failure of many P. aeruginosa biofilm-related infections.
Keywords: Biofilms; Cyclic-di-GMP; Experimental biofilm evolution; Pseudomonas aeruginosa; Sigma factor RpoS; Virulence.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Pseudomonas aeruginosa Interstrain Dynamics and Selection of Hyperbiofilm Mutants during a Chronic Infection.mBio. 2019 Aug 13;10(4):e01698-19. doi: 10.1128/mBio.01698-19. mBio. 2019. PMID: 31409682 Free PMC article.
-
Evolution of Antibiotic Resistance in Biofilm and Planktonic Pseudomonas aeruginosa Populations Exposed to Subinhibitory Levels of Ciprofloxacin.Antimicrob Agents Chemother. 2018 Jul 27;62(8):e00320-18. doi: 10.1128/AAC.00320-18. Print 2018 Aug. Antimicrob Agents Chemother. 2018. PMID: 29760140 Free PMC article.
-
Pseudomonas aeruginosa Biofilm Antibiotic Resistance Gene ndvB Expression Requires the RpoS Stationary-Phase Sigma Factor.Appl Environ Microbiol. 2018 Mar 19;84(7):e02762-17. doi: 10.1128/AEM.02762-17. Print 2018 Apr 1. Appl Environ Microbiol. 2018. PMID: 29352081 Free PMC article.
-
Thermoregulation of Pseudomonas aeruginosa Biofilm Formation.Appl Environ Microbiol. 2020 Oct 28;86(22):e01584-20. doi: 10.1128/AEM.01584-20. Print 2020 Oct 28. Appl Environ Microbiol. 2020. PMID: 32917757 Free PMC article.
-
c-di-GMP and its Effects on Biofilm Formation and Dispersion: a Pseudomonas Aeruginosa Review.Microbiol Spectr. 2015 Apr;3(2):MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014. Microbiol Spectr. 2015. PMID: 26104694 Free PMC article. Review.
Cited by
-
Divergent molecular strategies drive evolutionary adaptation to competitive fitness in biofilm formation.ISME J. 2024 Jan 8;18(1):wrae135. doi: 10.1093/ismejo/wrae135. ISME J. 2024. PMID: 39052320 Free PMC article.
-
Metabolomics responses and tolerance of Pseudomonas aeruginosa under acoustic vibration stress.PLoS One. 2024 Jan 29;19(1):e0297030. doi: 10.1371/journal.pone.0297030. eCollection 2024. PLoS One. 2024. PMID: 38285708 Free PMC article.
-
Modeling Polygenic Antibiotic Resistance Evolution in Biofilms.Front Microbiol. 2022 Jul 7;13:916035. doi: 10.3389/fmicb.2022.916035. eCollection 2022. Front Microbiol. 2022. PMID: 35875522 Free PMC article.
-
Glucose alters the evolutionary response to gentamicin in uropathogenic Escherichia coli.Microbiology (Reading). 2025 Mar;171(3):001548. doi: 10.1099/mic.0.001548. Microbiology (Reading). 2025. PMID: 40153309 Free PMC article.
-
Transcriptional Regulators Controlling Virulence in Pseudomonas aeruginosa.Int J Mol Sci. 2023 Jul 25;24(15):11895. doi: 10.3390/ijms241511895. Int J Mol Sci. 2023. PMID: 37569271 Free PMC article. Review.
References
-
- Duan X, Fu Y, Yang L. Molecular and systems biology approaches for analyzing drug-tolerant bacterial persister cells. In Sustainable Agriculture Reviews 46. Springer. 2020, pp 109–128.
Grants and funding
LinkOut - more resources
Full Text Sources

