3-hydroxy-L-kynurenamine is an immunomodulatory biogenic amine
- PMID: 34290243
- PMCID: PMC8295276
- DOI: 10.1038/s41467-021-24785-3
3-hydroxy-L-kynurenamine is an immunomodulatory biogenic amine
Abstract
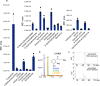
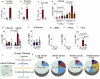
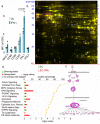
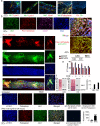
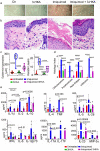
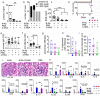
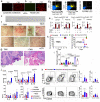
Tryptophan catabolism is a major metabolic pathway utilized by several professional and non-professional antigen presenting cells to maintain immunological tolerance. Here we report that 3-hydroxy-L-kynurenamine (3-HKA) is a biogenic amine produced via an alternative pathway of tryptophan metabolism. In vitro, 3-HKA has an anti-inflammatory profile by inhibiting the IFN-γ mediated STAT1/NF-κΒ pathway in both mouse and human dendritic cells (DCs) with a consequent decrease in the release of pro-inflammatory chemokines and cytokines, most notably TNF, IL-6, and IL12p70. 3-HKA has protective effects in an experimental mouse model of psoriasis by decreasing skin thickness, erythema, scaling and fissuring, reducing TNF, IL-1β, IFN-γ, and IL-17 production, and inhibiting generation of effector CD8+ T cells. Similarly, in a mouse model of nephrotoxic nephritis, besides reducing inflammatory cytokines, 3-HKA improves proteinuria and serum urea nitrogen, overall ameliorating immune-mediated glomerulonephritis and renal dysfunction. Overall, we propose that this biogenic amine is a crucial component of tryptophan-mediated immune tolerance.
© 2021. The Author(s).
Conflict of interest statement
L.S. declares that 3-HKA is covered by patent application # PCT/US2019/21731 to Albert Einstein College of Medicine. The other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

