Targeting the latent human cytomegalovirus reservoir for T-cell-mediated killing with virus-specific nanobodies
- PMID: 34290252
- PMCID: PMC8295288
- DOI: 10.1038/s41467-021-24608-5
Targeting the latent human cytomegalovirus reservoir for T-cell-mediated killing with virus-specific nanobodies
Abstract
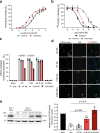
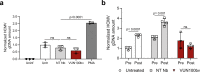
Latent human cytomegalovirus (HCMV) infection is characterized by limited gene expression, making latent HCMV infections refractory to current treatments targeting viral replication. However, reactivation of latent HCMV in immunosuppressed solid organ and stem cell transplant patients often results in morbidity. Here, we report the killing of latently infected cells via a virus-specific nanobody (VUN100bv) that partially inhibits signaling of the viral receptor US28. VUN100bv reactivates immediate early gene expression in latently infected cells without inducing virus production. This allows recognition and killing of latently infected monocytes by autologous cytotoxic T lymphocytes from HCMV-seropositive individuals, which could serve as a therapy to reduce the HCMV latent reservoir of transplant patients.
© 2021. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: R.H. is affiliated with QVQ Holding BV, a company offering VHH services and VHH-based imaging molecules. T.D.G, E.E., R.H., J.S., and M.S. are co-inventors on a pending EU patent application EP19190047.1, which covers the use of US28-targeting nanobodies for T-cell-mediated killing of HCMV latently infected cells. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

