Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma
- PMID: 34290406
- PMCID: PMC9187974
- DOI: 10.1038/s41586-021-03704-y
Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma
Abstract
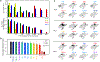
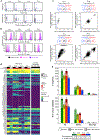
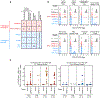
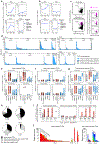
Interactions between T cell receptors (TCRs) and their cognate tumour antigens are central to antitumour immune responses1-3; however, the relationship between phenotypic characteristics and TCR properties is not well elucidated. Here we show, by linking the antigenic specificity of TCRs and the cellular phenotype of melanoma-infiltrating lymphocytes at single-cell resolution, that tumour specificity shapes the expression state of intratumoural CD8+ T cells. Non-tumour-reactive T cells were enriched for viral specificities and exhibited a non-exhausted memory phenotype, whereas melanoma-reactive lymphocytes predominantly displayed an exhausted state that encompassed diverse levels of differentiation but rarely acquired memory properties. These exhausted phenotypes were observed both among clonotypes specific for public overexpressed melanoma antigens (shared across different tumours) or personal neoantigens (specific for each tumour). The recognition of such tumour antigens was provided by TCRs with avidities inversely related to the abundance of cognate targets in melanoma cells and proportional to the binding affinity of peptide-human leukocyte antigen (HLA) complexes. The persistence of TCR clonotypes in peripheral blood was negatively affected by the level of intratumoural exhaustion, and increased in patients with a poor response to immune checkpoint blockade, consistent with chronic stimulation mediated by residual tumour antigens. By revealing how the quality and quantity of tumour antigens drive the features of T cell responses within the tumour microenvironment, we gain insights into the properties of the anti-melanoma TCR repertoire.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures










Comment in
-
Tumour antigen-induced T cell exhaustion - the archenemy of immune-hot malignancies.Nat Rev Clin Oncol. 2021 Dec;18(12):749-750. doi: 10.1038/s41571-021-00562-5. Nat Rev Clin Oncol. 2021. PMID: 34556846 Free PMC article.
-
Phenotypic characteristics and T cell receptor properties in melanoma: deciphering the correlation at single-cell resolution.Signal Transduct Target Ther. 2022 Jan 4;7(1):5. doi: 10.1038/s41392-021-00864-1. Signal Transduct Target Ther. 2022. PMID: 34983923 Free PMC article. No abstract available.
References
Methods references
-
- Haga-Friedman A, Horovitz-Fried M & Cohen CJ Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J Immunol 188, 5538–5546 (2012). - PubMed
-
- Bialer G, Horovitz-Fried M, Ya’acobi S, Morgan RA & Cohen CJ Selected murine residues endow human TCR with enhanced tumor recognition. J Immunol 184, 6232–6241 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- P01 CA229092/CA/NCI NIH HHS/United States
- R01 AI046223/AI/NIAID NIH HHS/United States
- P50 CA101942/CA/NCI NIH HHS/United States
- U19 AI082630/AI/NIAID NIH HHS/United States
- R01 CA155010/CA/NCI NIH HHS/United States
- R01 CA229261/CA/NCI NIH HHS/United States
- R01 CA279391/CA/NCI NIH HHS/United States
- R01 DK124336/DK/NIDDK NIH HHS/United States
- U24 CA224331/CA/NCI NIH HHS/United States
- R50 CA211482/CA/NCI NIH HHS/United States
- 5P30 CA042014-24/NH/NIH HHS/United States
- R21 CA216772/CA/NCI NIH HHS/United States
- R01 CA208756/CA/NCI NIH HHS/United States
- R50 CA251956/CA/NCI NIH HHS/United States
- R01 AG047956/AG/NIA NIH HHS/United States
- R01 AI141202/AI/NIAID NIH HHS/United States
- R00 HL102228-05/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

