Novel lncRNA lncRNA001074 participates in the low salinity-induced response in the sea cucumber Apostichopus japonicus by targeting the let-7/NKAα axis
- PMID: 34291427
- PMCID: PMC8492809
- DOI: 10.1007/s12192-021-01207-3
Novel lncRNA lncRNA001074 participates in the low salinity-induced response in the sea cucumber Apostichopus japonicus by targeting the let-7/NKAα axis
Abstract
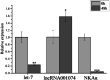
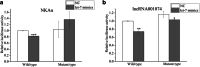
Salinity fluctuations have severe impacts on sea cucumbers and therefore important consequences in sea cucumber farming. The responses of sea cucumbers to salinity changes are reflected in the expression profiles of multiple genes and non-coding RNAs (ncRNAs). The microRNA (let-7) which is a developmental regulator, the ion transporter gene sodium potassium ATPase gene (NKAα), and the long ncRNA lncRNA001074 were previously shown to be involved in responses to salinity changes in various marine species. To better understand the relationship between ncRNAs and target genes, the let-7/NKAα/lncRNA001074 predicted interaction was investigated in this study using luciferase reporter assays and gene knockdowns in the sea cucumber Apostichopus japonicus. The results showed that NKAα was the target gene of let-7 and NKAα expression levels were inversely correlated with let-7 expression based on the luciferase reporter assays and western blots. The let-7 abundance was negatively regulated by lncRNA001074 and NKAα both in vitro and in vivo. Knockdown of lncRNA001074 led to let-7 overexpression. These results demonstrated that lncRNA001074 binds to the 3'-UTR binding site of let-7 in a regulatory manner. Furthermore, the expression profiles of let-7, NKAα, and lncRNA001074 were analyzed in sea cucumbers after the knockdown of each of these genes. The results found that lncRNA001074 competitively bound let-7 to suppress NKAα expression under low salinity conditions. The downregulation of let-7, in conjunction with the upregulation of lncRNA001074 and NKAα, may be essential for the response to low salinity change in sea cucumbers. Therefore, the dynamic balance of the lncRNA001074, NKAα, and let-7 network might be a potential response mechanism to salinity change in sea cucumbers.
Keywords: Apostichopus japonicus; Na+/K+-ATPase α; Salinity response; ceRNA; let-7; lncRNA001074.
© 2021. Cell Stress Society International.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Asha PS, Muthiah P. Effects of temperature, salinity and pH on larval growth, survival and development of the sea cucumber Holothuria spinifera Theel. Aquaculture. 2005;250:823–829. doi: 10.1016/j.aquaculture.2005.04.075. - DOI
-
- Binyon J. The effects of diluted sea water upon podial tissues of the starfish Asterias rubens L. Comp Biochem Physiol A Physiol. 1972;41:1–6. doi: 10.1016/0300-9629(72)90026-6. - DOI
-
- Bozza DC, Freire CA, Prodocimo V. Osmo-ionic regulation and carbonic anhydrase, Na/K-ATPase and V-H-ATPase activities in gills of the ancient freshwater crustacean Aegla schmitti (Anomura) exposed to high salinities Comparative biochemistry and physiology Part A. Mol Integr Physiol. 2019;231:201–208. doi: 10.1016/j.cbpa.2019.02.024. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

