Mutational analysis reveals potential phosphorylation sites in eukaryotic elongation factor 1A that are important for its activity
- PMID: 34293820
- PMCID: PMC9292714
- DOI: 10.1002/1873-3468.14164
Mutational analysis reveals potential phosphorylation sites in eukaryotic elongation factor 1A that are important for its activity
Abstract
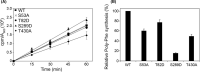
Previous studies have suggested that phosphorylation of translation elongation factor 1A (eEF1A) can alter its function, and large-scale phospho-proteomic analyses in Saccharomyces cerevisiae have identified 14 eEF1A residues phosphorylated under various conditions. Here, a series of eEF1A mutations at these proposed sites were created and the effects on eEF1A activity were analyzed. The eEF1A-S53D and eEF1A-T430D phosphomimetic mutant strains were inviable, while corresponding alanine mutants survived but displayed defects in growth and protein synthesis. The activity of an eEF1A-S289D mutant was significantly reduced in the absence of the guanine nucleotide exchange factor eEF1Bα and could be restored by an exchange-deficient form of the protein, suggesting that eEF1Bα promotes eEF1A activity by a mechanism other than nucleotide exchange. Our data show that several of the phosphorylation sites identified by high-throughput analysis are critical for eEF1A function.
Keywords: elongation factor; guanine nucleotide exchange factor; phosphorylation; translation; yeast.
© 2021 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures




Similar articles
-
Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Balpha.J Biol Chem. 2009 Feb 13;284(7):4739-47. doi: 10.1074/jbc.M807945200. Epub 2008 Dec 18. J Biol Chem. 2009. PMID: 19095653 Free PMC article.
-
Methylation of elongation factor 1A by yeast Efm4 or human eEF1A-KMT2 involves a beta-hairpin recognition motif and crosstalks with phosphorylation.J Biol Chem. 2024 Feb;300(2):105639. doi: 10.1016/j.jbc.2024.105639. Epub 2024 Jan 8. J Biol Chem. 2024. PMID: 38199565 Free PMC article.
-
Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange.J Biol Chem. 1999 Oct 15;274(42):30297-302. doi: 10.1074/jbc.274.42.30297. J Biol Chem. 1999. PMID: 10514524
-
Anticancer Small-Molecule Agents Targeting Eukaryotic Elongation Factor 1A: State of the Art.Int J Mol Sci. 2023 Mar 8;24(6):5184. doi: 10.3390/ijms24065184. Int J Mol Sci. 2023. PMID: 36982256 Free PMC article. Review.
-
Ser/Thr kinases and polyamines in the regulation of non-canonical functions of elongation factor 1A.Amino Acids. 2016 Oct;48(10):2339-52. doi: 10.1007/s00726-016-2311-3. Epub 2016 Aug 27. Amino Acids. 2016. PMID: 27568183 Review.
Cited by
-
eEF1α2 is required for actin cytoskeleton homeostasis in the aging muscle.Dis Model Mech. 2024 Sep 1;17(9):dmm050729. doi: 10.1242/dmm.050729. Epub 2024 Aug 29. Dis Model Mech. 2024. PMID: 39207054 Free PMC article.
-
Glycosylating Effectors of Legionella pneumophila: Finding the Sweet Spots for Host Cell Subversion.Biomolecules. 2022 Feb 4;12(2):255. doi: 10.3390/biom12020255. Biomolecules. 2022. PMID: 35204756 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

