METAMVGL: a multi-view graph-based metagenomic contig binning algorithm by integrating assembly and paired-end graphs
- PMID: 34294039
- PMCID: PMC8296540
- DOI: 10.1186/s12859-021-04284-4
METAMVGL: a multi-view graph-based metagenomic contig binning algorithm by integrating assembly and paired-end graphs
Abstract
Background: Due to the complexity of microbial communities, de novo assembly on next generation sequencing data is commonly unable to produce complete microbial genomes. Metagenome assembly binning becomes an essential step that could group the fragmented contigs into clusters to represent microbial genomes based on contigs' nucleotide compositions and read depths. These features work well on the long contigs, but are not stable for the short ones. Contigs can be linked by sequence overlap (assembly graph) or by the paired-end reads aligned to them (PE graph), where the linked contigs have high chance to be derived from the same clusters.
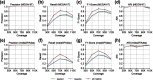
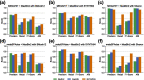
Results: We developed METAMVGL, a multi-view graph-based metagenomic contig binning algorithm by integrating both assembly and PE graphs. It could strikingly rescue the short contigs and correct the binning errors from dead ends. METAMVGL learns the two graphs' weights automatically and predicts the contig labels in a uniform multi-view label propagation framework. In experiments, we observed METAMVGL made use of significantly more high-confidence edges from the combined graph and linked dead ends to the main graph. It also outperformed many state-of-the-art contig binning algorithms, including MaxBin2, MetaBAT2, MyCC, CONCOCT, SolidBin and GraphBin on the metagenomic sequencing data from simulation, two mock communities and Sharon infant fecal samples.
Conclusions: Our findings demonstrate METAMVGL outstandingly improves the short contig binning and outperforms the other existing contig binning tools on the metagenomic sequencing data from simulation, mock communities and infant fecal samples.
Keywords: Assembly graph; Contig binning; Dead end; Multi-view label propagation; Paired-end graph.
© 2021. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest.
Figures


Similar articles
-
GraphBin: refined binning of metagenomic contigs using assembly graphs.Bioinformatics. 2020 Jun 1;36(11):3307-3313. doi: 10.1093/bioinformatics/btaa180. Bioinformatics. 2020. PMID: 32167528
-
Accurate Binning of Metagenomic Contigs Using Composition, Coverage, and Assembly Graphs.J Comput Biol. 2022 Dec;29(12):1357-1376. doi: 10.1089/cmb.2022.0262. Epub 2022 Nov 11. J Comput Biol. 2022. PMID: 36367700
-
MetaCluster-TA: taxonomic annotation for metagenomic data based on assembly-assisted binning.BMC Genomics. 2014;15 Suppl 1(Suppl 1):S12. doi: 10.1186/1471-2164-15-S1-S12. Epub 2014 Jan 24. BMC Genomics. 2014. PMID: 24564377 Free PMC article.
-
Application of computational approaches to analyze metagenomic data.J Microbiol. 2021 Mar;59(3):233-241. doi: 10.1007/s12275-021-0632-8. Epub 2021 Feb 10. J Microbiol. 2021. PMID: 33565054 Review.
-
Metagenomic tools in microbial ecology research.Curr Opin Biotechnol. 2021 Feb;67:184-191. doi: 10.1016/j.copbio.2021.01.019. Epub 2021 Feb 14. Curr Opin Biotechnol. 2021. PMID: 33592536 Review.
Cited by
-
Unitig level assembly graph based metagenome-assembled genome refiner (UGMAGrefiner): A tool to increase completeness and resolution of metagenome-assembled genomes.Comput Struct Biotechnol J. 2023 Mar 21;21:2394-2404. doi: 10.1016/j.csbj.2023.03.030. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37066122 Free PMC article.
-
Applications of de Bruijn graphs in microbiome research.Imeta. 2022 Mar 1;1(1):e4. doi: 10.1002/imt2.4. eCollection 2022 Mar. Imeta. 2022. PMID: 38867733 Free PMC article. Review.
-
Constructing metagenome-assembled genomes for almost all components in a real bacterial consortium for binning benchmarking.BMC Genomics. 2022 Nov 10;23(1):746. doi: 10.1186/s12864-022-08967-x. BMC Genomics. 2022. PMID: 36352370 Free PMC article.
-
A toolbox of machine learning software to support microbiome analysis.Front Microbiol. 2023 Nov 22;14:1250806. doi: 10.3389/fmicb.2023.1250806. eCollection 2023. Front Microbiol. 2023. PMID: 38075858 Free PMC article. Review.
-
A review of computational tools for generating metagenome-assembled genomes from metagenomic sequencing data.Comput Struct Biotechnol J. 2021 Nov 23;19:6301-6314. doi: 10.1016/j.csbj.2021.11.028. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34900140 Free PMC article. Review.
References
-
- Poyet M, Groussin M, Gibbons S, Avila-Pacheco J, Jiang X, Kearney S, Perrotta A, Berdy B, Zhao S, Lieberman T, et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat Med. 2019;25(9):1442–1452. doi: 10.1038/s41591-019-0559-3. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

