No evidence for differential gene expression in major depressive disorder PBMCs, but robust evidence of elevated biological ageing
- PMID: 34294682
- PMCID: PMC8298604
- DOI: 10.1038/s41398-021-01506-4
No evidence for differential gene expression in major depressive disorder PBMCs, but robust evidence of elevated biological ageing
Abstract
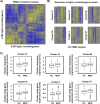
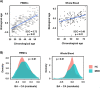
The increasingly compelling data supporting the involvement of immunobiological mechanisms in Major Depressive Disorder (MDD) might provide some explanation forthe variance in this heterogeneous condition. Peripheral blood measures of cytokines and chemokines constitute the bulk of evidence, with consistent meta-analytic data implicating raised proinflammatory cytokines such as IL6, IL1β and TNF. Among the potential mechanisms linking immunobiological changes to affective neurobiology is the accelerated biological ageing seen in MDD, particularly via the senescence associated secretory phenotype (SASP). However, the cellular source of immunobiological markers remains unclear. Pre-clinical evidence suggests a role for peripheral blood mononuclear cells (PBMC), thus here we aimed to explore the transcriptomic profile using RNA sequencing in PBMCs in a clinical sample of people with various levels of depression and treatment response comparing it with that in healthy controls (HCs). There were three groups with major depressive disorder (MDD): treatment-resistant (n = 94), treatment-responsive (n = 47) and untreated (n = 46). Healthy controls numbered 44. Using PBMCs gene expression analysis was conducted using RNAseq to a depth of 54.5 million reads. Differential gene expression analysis was performed using DESeq2. The data showed no robust signal differentiating MDD and HCs. There was, however, significant evidence of elevated biological ageing in MDD vs HC. Biological ageing was evident in these data as a transcriptional signature of 888 age-associated genes (adjusted p < 0.05, absolute log2fold > 0.6) that also correlated strongly with chronological age (spearman correlation coefficient of 0.72). Future work should expand clinical sample sizes and reduce clinical heterogeneity. Exploration of RNA-seq signatures in other leukocyte populations and single cell RNA sequencing may help uncover more subtle differences. However, currently the subtlety of any PBMC signature mitigates against its convincing use as a diagnostic or predictive biomarker.
© 2021. The Author(s).
Conflict of interest statement
This work was funded by a Wellcome Trust strategy award to the Neuroimmunology of Mood Disorders and Alzheimer’s Disease (NIMA) Consortium, which is also funded by Janssen, GlaxoSmithKline, Lundbeck and Pfizer. Dr. Drevets and Dr. De Boer are employees of Janssen Research & Development, LLC, of Johnson & Johnson and hold equity in Johnson & Johnson.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

