CD8+ tissue-resident memory T cells promote liver fibrosis resolution by inducing apoptosis of hepatic stellate cells
- PMID: 34294714
- PMCID: PMC8298513
- DOI: 10.1038/s41467-021-24734-0
CD8+ tissue-resident memory T cells promote liver fibrosis resolution by inducing apoptosis of hepatic stellate cells
Abstract
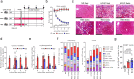
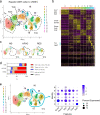
Non-alcoholic steatohepatitis (NASH) is a leading cause of chronic liver disease that can progress to liver fibrosis. Recent clinical advance suggests a reversibility of liver fibrosis, but the cellular and molecular mechanisms underlying NASH resolution remain unclarified. Here, using a murine diet-induced NASH and the subsequent resolution model, we demonstrate direct roles of CD8+ tissue-resident memory CD8+ T (CD8+ Trm) cells in resolving liver fibrosis. Single-cell transcriptome analysis and FACS analysis revealed CD69+CD103-CD8+ Trm cell enrichment in NASH resolution livers. The reduction of liver CD8+ Trm cells, maintained by tissue IL-15, significantly delayed fibrosis resolution, while adoptive transfer of these cells protected mice from fibrosis progression. During resolution, CD8+ Trm cells attracted hepatic stellate cells (HSCs) in a CCR5-dependent manner, and predisposed activated HSCs to FasL-Fas-mediated apoptosis. Histological assessment of patients with NASH revealed CD69+CD8+ Trm abundance in fibrotic areas, further supporting their roles in humans. These results highlight the undefined role of liver CD8+ Trm in fibrosis resolution.
© 2021. The Author(s).
Conflict of interest statement
This study was funded in part by Mitsubishi Tanabe Pharma Corporation. Y.K. is an employee of Mitsubishi Tanabe Pharma Corporation. The remaining authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

