Impaired SNF2L Chromatin Remodeling Prolongs Accessibility at Promoters Enriched for Fos/Jun Binding Sites and Delays Granule Neuron Differentiation
- PMID: 34295220
- PMCID: PMC8290069
- DOI: 10.3389/fnmol.2021.680280
Impaired SNF2L Chromatin Remodeling Prolongs Accessibility at Promoters Enriched for Fos/Jun Binding Sites and Delays Granule Neuron Differentiation
Abstract
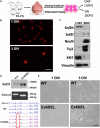
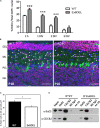
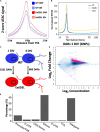
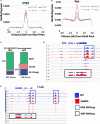
Chromatin remodeling proteins utilize the energy from ATP hydrolysis to mobilize nucleosomes often creating accessibility for transcription factors within gene regulatory elements. Aberrant chromatin remodeling has diverse effects on neuroprogenitor homeostasis altering progenitor competence, proliferation, survival, or cell fate. Previous work has shown that inactivation of the ISWI genes, Smarca5 (encoding Snf2h) and Smarca1 (encoding Snf2l) have dramatic effects on brain development. Smarca5 conditional knockout mice have reduced progenitor expansion and severe forebrain hypoplasia, with a similar effect on the postnatal growth of the cerebellum. In contrast, Smarca1 mutants exhibited enlarged forebrains with delayed progenitor differentiation and increased neuronal output. Here, we utilized cerebellar granule neuron precursor (GNP) cultures from Smarca1 mutant mice (Ex6DEL) to explore the requirement for Snf2l on progenitor homeostasis. The Ex6DEL GNPs showed delayed differentiation upon plating that was not attributed to changes in the Sonic Hedgehog pathway but was associated with overexpression of numerous positive effectors of proliferation, including targets of Wnt activation. Transcriptome analysis identified increased expression of Fosb and Fosl2 while ATACseq experiments identified a large increase in chromatin accessibility at promoters many enriched for Fos/Jun binding sites. Nonetheless, the elevated proliferation index was transient and the Ex6DEL cultures initiated differentiation with a high concordance in gene expression changes to the wild type cultures. Genes specific to Ex6DEL differentiation were associated with an increased activation of the ERK signaling pathway. Taken together, this data provides the first indication of how Smarca1 mutations alter progenitor cell homeostasis and contribute to changes in brain size.
Keywords: ATAC-seq; ISWI chromatin remodeler; SMARCA1; Snf2L; cerebellar granule neuron progenitors; chromatin accessibility.
Copyright © 2021 Goodwin, Zapata, Timpano, Marenger and Picketts.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

