Extracellular vesicle miRNA predict FDG-PET status in patients with classical Hodgkin Lymphoma
- PMID: 34295456
- PMCID: PMC8282992
- DOI: 10.1002/jev2.12121
Extracellular vesicle miRNA predict FDG-PET status in patients with classical Hodgkin Lymphoma
Abstract
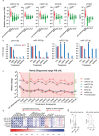
Minimally-invasive tools to assess tumour presence and burden may improve clinical management. FDG-PET (metabolic) imaging is the current gold standard for interim response assessment in patients with classical Hodgkin Lymphoma (cHL), but this technique cannot be repeated frequently. Here we show that microRNAs (miRNA) associated with tumour-secreted extracellular vesicles (EVs) in the circulation of cHL patients may improve response assessment. Small RNA sequencing and qRT-PCR reveal that the relative abundance of cHL-expressed miRNAs, miR-127-3p, miR-155-5p, miR-21-5p, miR-24-3p and let-7a-5p is up to hundred-fold increased in plasma EVs of cHL patients pre-treatment when compared to complete metabolic responders (CMR). Notably, in partial responders (PR) or treatment-refractory cases (n = 10) the EV-miRNA levels remain elevated. In comparison, tumour specific copy number variations (CNV) were detected in cell-free DNA of 8 out of 10 newly diagnosed cHL patients but not in patients with PR. Combining EV-miR-127-3p and/or EV-let-7a-5p levels, with serum TARC (a validated protein cHL biomarker), increases the accuracy for predicting PET-status (n = 129) to an area under the curve of 0.93 (CI: 0.87-0.99), 93.5% sensitivity, 83.8/85.0% specificity and a negative predictive value of 96%. Thus the level of tumour-associated miRNAs in plasma EVs is predictive of metabolic tumour activity in cHL patients. Our findings suggest that plasma EV-miRNA are useful for detection of small residual lesions and may be applied as serial response prediction tool.
Keywords: Hodgkin lymphoma; blood; extracellular vesicles; liquid biopsy; miRNA; response monitoring.
© 2021 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Conflict of interest statement
Dirk Michiel Pegtel and Michael Hackenberg are co‐founders of Exbiome BV. Dirk Michiel Pegtel is CSO of ExBiome BV and has received travel compensation from Takeda.
Figures



References
-
- Adams, H. J. A. , Nievelstein, R. A. J. , & Kwee, T. C. (2015). Prognostic value of interim FDG‐PET in Hodgkin lymphoma: Systematic review and meta‐analysis. British Journal of Haematology, 170(3), 356–366. - PubMed
-
- Amant, F. , Verheecke, M. , Wlodarska, I. , Dehaspe, L. , Brady, P. , Brison, N. , Van Den Bogaert, K. , Dierickx, D. , Vandecaveye, V. , Tousseyn, T. , Moerman, P. , Vanderstichele, A. , Vergote, I. , Neven, P. , Berteloot, P. , Putseys, K. , Danneels, L. , Vandenberghe, P. , Legius, E. , & Vermeesch, J. R. (2015). Presymptomatic Identification of Cancers in Pregnant Women During Noninvasive Prenatal Testing. JAMA Oncology, 1(6), 814–819. 10.1001/jamaoncol.2015.1883. - DOI - PubMed
-
- Anfossi, S. , Babayan, A. , Pantel, K. , & Calin, G. A. (2018). Clinical utility of circulating non‐coding RNAs — an update. Nature Reviews Clinical Oncology, 15(9), 541–563. - PubMed
-
- Aparicio‐Puerta, E. , Lebrón, R. , Rueda, A. , Gómez‐Martín, C. , Giannoukakos, S. , Jaspez, D. , Medina, J. M. , Zubkovic, A. , Jurak, I. , Fromm, B. , Marchal, J. A. , Oliver, J. , & Hackenberg, M. (2019). sRNAbench and sRNAtoolbox 2019: Intuitive fast small RNA profiling and differential expression. Nucleic Acids Research, 47(W1), W530–W535. - PMC - PubMed
-
- Barturen, G. , Rueda, A. , Hamberg, M. , Alganza, A. , Lebron, R. , Kotsyfakis, M. , Shi, B.‐J. , Koppers‐Lalic, D. , & Hackenberg, M. (2014). sRNAbench: profiling of small RNAs and its sequence variants in single or multi‐species high‐throughput experiments. Methods in Next Generation Sequencing, 1(1), 10.2478/mngs-2014-0001. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

