Identification of differentially expressed proteins in the locoregional recurrent esophageal squamous cell carcinoma by quantitative proteomics
- PMID: 34295551
- PMCID: PMC8261328
- DOI: 10.21037/jgo-21-278
Identification of differentially expressed proteins in the locoregional recurrent esophageal squamous cell carcinoma by quantitative proteomics
Abstract
Background: This study aimed to identify potential biomarkers associated with locoregional recurrence in patients with esophageal squamous cell carcinoma (ESCC) after radical resection.
Methods: We performed a quantitative proteomics analysis using isobaric tags for relative and absolute quantification (iTRAQ) with reversed-phase liquid chromatography-mass spectrometry (RPLC-MS) to identify differential expression proteins (DEPs) between a locoregional recurrence group and good prognosis group of ESCC after radical esophagectomy. The bioinformatics analysis was performed with ingenuity pathway analysis software (IPA) and Gene Ontology (GO) database using the software of MAS 3.0. Kaplan-Meier (KM) Plotter Online Tool (http://www.kmplot.com) was used to evaluate the relationship between the differential expression of proteins and survival in patients with ESCC.
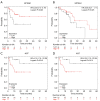
Results: More than 400 proteins were quantitated of which 27 proteins had upregulated expression and 55 proteins had downregulated expression in the locoregional recurrence group compared to the good prognosis group. These 82 DEPs were associated with biological procession of cancer development including cellular movement, cellular assembly and organization, cellular function and maintenance, cellular growth and proliferation, cell death and survival, DNA replication recombination and repair, and so on. Of these DEPs, SPTAN1 and AGT proteins were identified to be associated with RFS in ESCC. SPTAN1 was positively associated with RFS and AGT was negatively associated with RFS. Expression of SPTAN1 tended to have favorable OS while expression of AGT tended to have poor OS.
Conclusions: Our results demonstrated that quantitative proteomics is an effective discovery tool to identify biomarkers for prognosis prediction in ESCC. However, it needs more studies with large populations of ESCC to validate these potential biomarkers.
Keywords: Biomarkers; esophageal carcinoma; quantitative proteomics; recurrence.
2021 Journal of Gastrointestinal Oncology. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-278). The authors have no conflicts of interest to declare.
Figures



Similar articles
-
Identification of extracellular matrix protein 1 as a potential plasma biomarker of ESCC by proteomic analysis using iTRAQ and 2D-LC-MS/MS.Proteomics Clin Appl. 2017 Sep;11(9-10). doi: 10.1002/prca.201600163. Epub 2017 May 29. Proteomics Clin Appl. 2017. PMID: 28493612
-
iTRAQ-based quantitative proteomic analysis of esophageal squamous cell carcinoma.Tumour Biol. 2016 Feb;37(2):1909-18. doi: 10.1007/s13277-015-3840-1. Epub 2015 Sep 2. Tumour Biol. 2016. PMID: 26330293
-
iTRAQ-Based Quantitative Proteomic Analyses of High Grade Esophageal Squamous Intraepithelial Neoplasia.Proteomics Clin Appl. 2017 Dec;11(11-12). doi: 10.1002/prca.201600167. Epub 2017 Sep 25. Proteomics Clin Appl. 2017. PMID: 28816019
-
Comparative proteome analysis of form-deprivation myopia in sclera with iTRAQ-based quantitative proteomics.Mol Vis. 2021 Sep 1;27:494-505. eCollection 2021. Mol Vis. 2021. PMID: 34526757 Free PMC article.
-
Identification of prothymosin alpha (PTMA) as a biomarker for esophageal squamous cell carcinoma (ESCC) by label-free quantitative proteomics and Quantitative Dot Blot (QDB).Clin Proteomics. 2019 Apr 5;16:12. doi: 10.1186/s12014-019-9232-6. eCollection 2019. Clin Proteomics. 2019. PMID: 30988666 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
