Gucy2d selectively marks inhibitory dynorphin neurons in the spinal dorsal horn but is dispensable for pain and itch sensitivity
- PMID: 34296052
- PMCID: PMC8291471
- DOI: 10.1097/PR9.0000000000000947
Gucy2d selectively marks inhibitory dynorphin neurons in the spinal dorsal horn but is dispensable for pain and itch sensitivity
Abstract
Introduction: Inhibitory neurons in the spinal dorsal horn can be classified based on expression of neurochemical marker genes. However, these marker genes are often expressed throughout the central nervous system, which poses challenges for manipulating genetically identified spinal neurons without undesired off-target effects.
Objectives: We investigated whether Gucy2d, previously identified as a highly selective marker of dynorphin-lineage neurons in the dorsal horn, is expressed in other locations within the adult mouse spinal cord, dorsal root ganglia (DRG), or brain. In addition, we sought to molecularly characterize Gucy2d-expressing dorsal horn neurons and investigate whether the disruption of Gucy2d gene expression affects sensitivity to itch or pain.
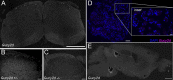
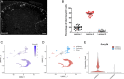
Methods: In situ hybridization experiments assessed Gucy2d mRNA expression in the adult mouse spinal cord, DRG, and brain, and its colocalization with Pax2, Bhlhb5, and Pde2a in dorsal horn neurons. Knockout mice lacking Gucy2d expression were compared with littermate controls to assess sensitivity to chloroquine-induced itch and dry skin-mediated chronic itch, as well as heat, cold, or mechanical stimuli.
Results: Gucy2d is selectively expressed in dynorphin-lineage neurons in lamina I-III of the adult mouse spinal cord but not in the brain or DRG. Spinal Gucy2d-expressing neurons are inhibitory neurons that also express the transcription factor Bhlhb5 and the cGMP-dependent phosphodiesterase Pde2a. Gucy2d knockout mice did not exhibit altered responses to itch or pain.
Conclusions: The selective expression of Gucy2d within a subpopulation of inhibitory dorsal horn neurons may yield a means to selectively manipulate inhibitory signaling at the level of the spinal cord without effects on the brain.
Keywords: Dynorphin; Guanylate cyclase; Gucy2d; Itch; Pain; Spinal cord.
Copyright © 2021 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of The International Association for the Study of Pain.
Conflict of interest statement
The authors have no conflicts of interest to declare.Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Figures




Similar articles
-
Development and characterization of a Gucy2d-cre mouse to selectively manipulate a subset of inhibitory spinal dorsal horn interneurons.PLoS One. 2024 Mar 14;19(3):e0300282. doi: 10.1371/journal.pone.0300282. eCollection 2024. PLoS One. 2024. PMID: 38483883 Free PMC article.
-
Neonatal Injury Evokes Persistent Deficits in Dynorphin Inhibitory Circuits within the Adult Mouse Superficial Dorsal Horn.J Neurosci. 2020 May 13;40(20):3882-3895. doi: 10.1523/JNEUROSCI.0029-20.2020. Epub 2020 Apr 14. J Neurosci. 2020. PMID: 32291327 Free PMC article.
-
The majority of dorsal spinal cord gastrin releasing peptide is synthesized locally whereas neuromedin B is highly expressed in pain- and itch-sensing somatosensory neurons.Mol Pain. 2012 Jul 9;8:52. doi: 10.1186/1744-8069-8-52. Mol Pain. 2012. PMID: 22776446 Free PMC article.
-
Transcriptional profile of spinal dynorphin-lineage interneurons in the developing mouse.Pain. 2019 Oct;160(10):2380-2397. doi: 10.1097/j.pain.0000000000001636. Pain. 2019. PMID: 31166300 Free PMC article.
-
Spinal Microcircuits and the Regulation of Itch.In: Carstens E, Akiyama T, editors. Itch: Mechanisms and Treatment. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. Chapter 20. In: Carstens E, Akiyama T, editors. Itch: Mechanisms and Treatment. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. Chapter 20. PMID: 24830016 Free Books & Documents. Review.
Cited by
-
Molecular Anatomy of Synaptic and Extrasynaptic Neurotransmission Between Nociceptive Primary Afferents and Spinal Dorsal Horn Neurons.Int J Mol Sci. 2025 Mar 6;26(5):2356. doi: 10.3390/ijms26052356. Int J Mol Sci. 2025. PMID: 40076973 Free PMC article. Review.
-
Progress in spinal cord organoid research: advancing understanding of neural development, disease modelling, and regenerative medicine.Biomater Transl. 2024 Nov 15;5(4):355-371. doi: 10.12336/biomatertransl.2024.04.003. eCollection 2024. Biomater Transl. 2024. PMID: 39872925 Free PMC article. Review.
-
Development and characterization of a Gucy2d-cre mouse to selectively manipulate a subset of inhibitory spinal dorsal horn interneurons.PLoS One. 2024 Mar 14;19(3):e0300282. doi: 10.1371/journal.pone.0300282. eCollection 2024. PLoS One. 2024. PMID: 38483883 Free PMC article.
-
Selective Involvement of a Subset of Spinal Dorsal Horn Neurons Operated by a Prodynorphin Promoter in Aβ Fiber-Mediated Neuropathic Allodynia-Like Behavioral Responses in Rats.Front Mol Neurosci. 2022 Jun 23;15:911122. doi: 10.3389/fnmol.2022.911122. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35813063 Free PMC article.
References
-
- Bernard JF, Dallel R, Raboisson P, Villanueva L, Le Bars D. Organization of the efferent projections from the spinal cervical enlargement to the parabrachial area and periaqueductal gray: a PHA-L study in the rat. J Comp Neurol 1995;353:480–505. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
