Compartmentalization-aided interaction screening reveals extensive high-order complexes within the SARS-CoV-2 proteome
- PMID: 34297909
- PMCID: PMC8285250
- DOI: 10.1016/j.celrep.2021.109482
Compartmentalization-aided interaction screening reveals extensive high-order complexes within the SARS-CoV-2 proteome
Erratum in
-
Compartmentalization-aided interaction screening reveals extensive high-order complexes within the SARS-CoV-2 proteome.Cell Rep. 2021 Oct 19;37(3):109778. doi: 10.1016/j.celrep.2021.109778. Cell Rep. 2021. PMID: 34686343 Free PMC article. No abstract available.
Abstract
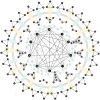
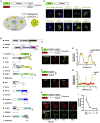
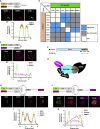
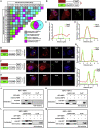
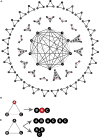
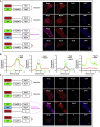
Bearing a relatively large single-stranded RNA genome in nature, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) utilizes sophisticated replication/transcription complexes (RTCs), mainly composed of a network of nonstructural proteins and nucleocapsid protein, to establish efficient infection. In this study, we develop an innovative interaction screening strategy based on phase separation in cellulo, namely compartmentalization of protein-protein interactions in cells (CoPIC). Utilizing CoPIC screening, we map the interaction network among RTC-related viral proteins. We identify a total of 47 binary interactions among 14 proteins governing replication, discontinuous transcription, and translation of coronaviruses. Further exploration via CoPIC leads to the discovery of extensive ternary complexes composed of these components, which infer potential higher-order complexes. Taken together, our results present an efficient and robust interaction screening strategy, and they indicate the existence of a complex interaction network among RTC-related factors, thus opening up opportunities to understand SARS-CoV-2 biology and develop therapeutic interventions for COVID-19.
Keywords: CoPIC; PRC2; RTC; SARS-CoV-2; high-order complexes; nonstructural proteins; protein-protein interaction.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Similar articles
-
SARS-CoV-2 Nonstructural Protein 1 Inhibits the Interferon Response by Causing Depletion of Key Host Signaling Factors.J Virol. 2021 Jun 10;95(13):e0026621. doi: 10.1128/JVI.00266-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 34110264 Free PMC article.
-
SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro.PLoS Pathog. 2008 May 2;4(5):e1000054. doi: 10.1371/journal.ppat.1000054. PLoS Pathog. 2008. PMID: 18451981 Free PMC article.
-
Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle.J Virol. 2020 Jan 31;94(4):e01925-19. doi: 10.1128/JVI.01925-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776274 Free PMC article.
-
Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19.Cells. 2021 Apr 6;10(4):821. doi: 10.3390/cells10040821. Cells. 2021. PMID: 33917481 Free PMC article. Review.
-
Overview of SARS-CoV-2 genome-encoded proteins.Sci China Life Sci. 2022 Feb;65(2):280-294. doi: 10.1007/s11427-021-1964-4. Epub 2021 Aug 10. Sci China Life Sci. 2022. PMID: 34387838 Free PMC article. Review.
Cited by
-
An efficient chemical screening method for structure-based inhibitors to nucleic acid enzymes targeting the DNA repair-replication interface and SARS CoV-2.Methods Enzymol. 2021;661:407-431. doi: 10.1016/bs.mie.2021.09.003. Epub 2021 Sep 27. Methods Enzymol. 2021. PMID: 34776222 Free PMC article.
-
Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.Nucleic Acids Res. 2022 Aug 12;50(14):8290-8301. doi: 10.1093/nar/gkac589. Nucleic Acids Res. 2022. PMID: 35801916 Free PMC article.
-
An atomistic model of the coronavirus replication-transcription complex as a hexamer assembled around nsp15.J Biol Chem. 2021 Oct;297(4):101218. doi: 10.1016/j.jbc.2021.101218. Epub 2021 Sep 23. J Biol Chem. 2021. PMID: 34562452 Free PMC article.
-
Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.bioRxiv [Preprint]. 2023 Nov 20:2023.11.17.567629. doi: 10.1101/2023.11.17.567629. bioRxiv. 2023. Update in: Protein Cell. 2024 Jul 1;15(7):547-552. doi: 10.1093/procel/pwae009. PMID: 38045375 Free PMC article. Updated. Preprint.
-
Flipped Over U: Structural Basis for dsRNA Cleavage by the SARS-CoV-2 Endoribonuclease.bioRxiv [Preprint]. 2022 Mar 2:2022.03.02.480688. doi: 10.1101/2022.03.02.480688. bioRxiv. 2022. Update in: Nucleic Acids Res. 2022 Aug 12;50(14):8290-8301. doi: 10.1093/nar/gkac589. PMID: 35262076 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

