Dual-AAV delivering split prime editor system for in vivo genome editing
- PMID: 34298129
- PMCID: PMC8753371
- DOI: 10.1016/j.ymthe.2021.07.011
Dual-AAV delivering split prime editor system for in vivo genome editing
Abstract
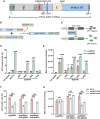
Prime editor (PE), a new genome editing tool, can generate all 12 possible base-to-base conversions, insertion, and deletion of short fragment DNA. PE has the potential to correct the majority of known human genetic disease-related mutations. Adeno-associated viruses (AAVs), the safe vector widely used in clinics, are not capable of delivering PE (∼6.3 kb) in a single vector because of the limited loading capacity (∼4.8 kb). To accommodate the loading capacity of AAVs, we constructed four split-PE (split-PE994, split-PE1005, split-PE1024, and split-PE1032) using Rma intein (Rhodothermus marinus). With the use of a GFP-mutated reporter system, PE reconstituting activities were screened, and two efficient split-PEs (split-PE1005 and split-PE1024) were identified. We then demonstrated that split-PEs delivered by dual-AAV1, especially split-PE1024, could mediate base transversion and insertion at four endogenous sites in human cells. To test the performance of split-PE in vivo, split-PE1024 was then delivered into the adult mouse retina by dual-AAV8. We demonstrated successful editing of Dnmt1 in adult mouse retina. Our study provides a new method to deliver PE to adult tissue, paving the way for in vivo gene-editing therapy using PE.
Keywords: dual-AAV; gene editing; in vivo; prime editor; split.
Copyright © 2021 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

