NPM1 Mutational Status Underlines Different Biological Features in Pediatric AML
- PMID: 34298672
- PMCID: PMC8304368
- DOI: 10.3390/cancers13143457
NPM1 Mutational Status Underlines Different Biological Features in Pediatric AML
Abstract
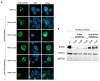
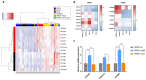
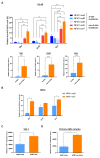
Nucleophosmin (NPM1) is a nucleocytoplasmic shuttling protein, predominantly located in the nucleolus, that regulates a multiplicity of different biological processes. NPM1 localization in the cell is finely tuned by specific signal motifs, with two tryptophan residues (Trp) being essential for the nucleolar localization. In acute myeloid leukemia (AML), several NPM1 mutations have been reported, all resulting in cytoplasmic delocalization, but the putative biological and clinical significance of different variants are still debated. We explored HOXA and HOXB gene expression profile in AML patients and found a differential expression between NPM1 mutations inducing the loss of two (A-like) Trp residues and those determining the loss of one Trp residue (non-A-like). We thus expressed NPM1 A-like- or non-A-like-mutated vectors in AML cell lines finding that NPM1 partially remained in the nucleolus in the non-A-like NPM1-mutated cells. As a result, only in A-like-mutated cells we detected HOXA5, HOXA10, and HOXB5 hyper-expression and p14ARF/p21/p53 pathway deregulation, leading to reduced sensitivity to the treatment with either chemotherapy or Venetoclax, as compared to non-A-like cells. Overall, we identified that the NPM1 mutational status mediates crucial biological characteristics of AML cells, providing the basis for further sub-classification and, potentially, management of this subgroup of patients.
Keywords: HOX genes; Nucleophosmin, NPM1; TP53; acute myeloid leukemia; drug treatment; gene expression; genetic; mutation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

