Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L
- PMID: 34299014
- PMCID: PMC8303626
- DOI: 10.3390/ijms22147396
Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L
Abstract
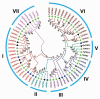
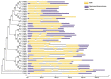
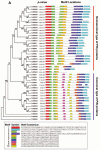
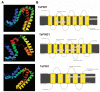
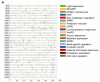
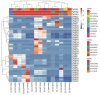
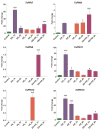
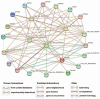
PIN-FORMED (PIN) genes play a crucial role in regulating polar auxin distribution in diverse developmental processes, including tropic responses, embryogenesis, tissue differentiation, and organogenesis. However, the role of PIN-mediated auxin transport in various plant species is poorly understood. Currently, no information is available about this gene family in wheat (Triticum aestivum L.). In the present investigation, we identified the PIN gene family in wheat to understand the evolution of PIN-mediated auxin transport and its role in various developmental processes and under different biotic and abiotic stress conditions. In this study, we performed genome-wide analysis of the PIN gene family in common wheat and identified 44 TaPIN genes through a homology search, further characterizing them to understand their structure, function, and distribution across various tissues. Phylogenetic analyses led to the classification of TaPIN genes into seven different groups, providing evidence of an evolutionary relationship with Arabidopsis thaliana and Oryza sativa. A gene exon/intron structure analysis showed a distinct evolutionary path and predicted the possible gene duplication events. Further, the physical and biochemical properties, conserved motifs, chromosomal, subcellular localization, transmembrane domains, and three-dimensional (3D) structure were also examined using various computational approaches. Cis-elements analysis of TaPIN genes showed that TaPIN promoters consist of phytohormone, plant growth and development, and stress-related cis-elements. In addition, expression profile analysis also revealed that the expression patterns of the TaPIN genes were different in different tissues and developmental stages. Several members of the TaPIN family were induced during biotic and abiotic stress. Moreover, the expression patterns of TaPIN genes were verified by qRT-PCR. The qRT-PCR results also show a similar expression with slight variation. Therefore, the outcome of this study provides basic genomic information on the expression of the TaPIN gene family and will pave the way for dissecting the precise role of TaPINs in plant developmental processes and different stress conditions.
Keywords: PIN; auxin; biotic and abiotic stress; cis-acting regulatory elements; polar auxin transport; qRT-PCR.
Conflict of interest statement
Authors declare no conflict of interest.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

