Antifungal Activity and Biosynthetic Potential of New Streptomyces sp. MW-W600-10 Strain Isolated from Coal Mine Water
- PMID: 34299061
- PMCID: PMC8303363
- DOI: 10.3390/ijms22147441
Antifungal Activity and Biosynthetic Potential of New Streptomyces sp. MW-W600-10 Strain Isolated from Coal Mine Water
Abstract
Crop infections by fungi lead to severe losses in food production and pose risks for human health. The increasing resistance of pathogens to fungicides has led to the higher usage of these chemicals, which burdens the environment and highlights the need to find novel natural biocontrol agents. Members of the genus Streptomyces are known to produce a plethora of bioactive compounds. Recently, researchers have turned to extreme and previously unexplored niches in the search for new strains with antimicrobial activities. One such niche are underground coal mine environments. We isolated the new Streptomyces sp. MW-W600-10 strain from coal mine water samples collected at 665 m below ground level. We examined the antifungal activity of the strain against plant pathogens Fusarium culmorum DSM62188 and Nigrospora oryzae roseF7. Furthermore, we analyzed the strain's biosynthetic potential with the antiSMASH tool. The strain showed inhibitory activity against both fungi strains. Genome mining revealed that it has 39 BGCs, among which 13 did not show similarity to those in databases. Additionally, we examined the activity of the Streptomyces sp. S-2 strain isolated from black soot against F. culmorum DSM62188. These results show that coal-related strains could be a source of novel bioactive compounds. Future studies will elucidate their full biotechnological potential.
Keywords: Streptomyces; antifungal activity; biosynthetic gene clusters; coal-related environments; gene expression; genome mining; quantitative PCR.
Conflict of interest statement
The authors do not have any conflict of interest.
Figures



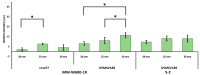
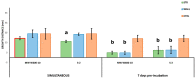

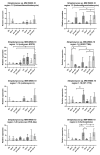
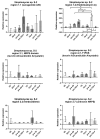
References
-
- Feng J., Hwang R., Chang K.F., Hwang S.F., Strelkov S.E., Gossen B.D., Conner R.L., Turnbull G.D. Genetic variation in Fusarium avenaceum causing root rot on field pea. Plant Pathol. 2010;59:845–852. doi: 10.1111/j.1365-3059.2010.02313.x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

