Longitudinal linked-read sequencing reveals ecological and evolutionary responses of a human gut microbiome during antibiotic treatment
- PMID: 34301627
- PMCID: PMC8327913
- DOI: 10.1101/gr.265058.120
Longitudinal linked-read sequencing reveals ecological and evolutionary responses of a human gut microbiome during antibiotic treatment
Abstract
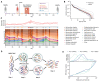
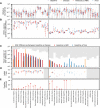
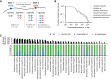
Gut microbial communities can respond to antibiotic perturbations by rapidly altering their taxonomic and functional composition. However, little is known about the strain-level processes that drive this collective response. Here, we characterize the gut microbiome of a single individual at high temporal and genetic resolution through a period of health, disease, antibiotic treatment, and recovery. We used deep, linked-read metagenomic sequencing to track the longitudinal trajectories of thousands of single nucleotide variants within 36 species, which allowed us to contrast these genetic dynamics with the ecological fluctuations at the species level. We found that antibiotics can drive rapid shifts in the genetic composition of individual species, often involving incomplete genome-wide sweeps of pre-existing variants. These genetic changes were frequently observed in species without obvious changes in species abundance, emphasizing the importance of monitoring diversity below the species level. We also found that many sweeping variants quickly reverted to their baseline levels once antibiotic treatment had concluded, demonstrating that the ecological resilience of the microbiota can sometimes extend all the way down to the genetic level. Our results provide new insights into the population genetic forces that shape individual microbiomes on therapeutically relevant timescales, with potential implications for personalized health and disease.
© 2021 Roodgar et al.; Published by Cold Spring Harbor Laboratory Press.
Figures