Pax5 regulates B cell immunity by promoting PI3K signaling via PTEN down-regulation
- PMID: 34301800
- PMCID: PMC7611449
- DOI: 10.1126/sciimmunol.abg5003
Pax5 regulates B cell immunity by promoting PI3K signaling via PTEN down-regulation
Abstract
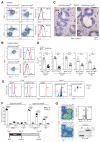
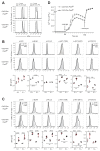
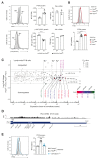
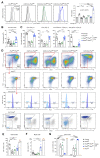
The transcription factor Pax5 controls B cell development, but its role in mature B cells is largely enigmatic. Here, we demonstrated that the loss of Pax5 by conditional mutagenesis in peripheral B lymphocytes led to the strong reduction of B-1a, marginal zone (MZ), and germinal center (GC) B cells as well as plasma cells. Follicular (FO) B cells tolerated the loss of Pax5 but had a shortened half-life. The Pax5-deficient FO B cells failed to proliferate upon B cell receptor or Toll-like receptor stimulation due to impaired PI3K-AKT signaling, which was caused by increased expression of PTEN, a negative regulator of the PI3K pathway. Pax5 restrained PTEN protein expression at the posttranscriptional level, likely involving Pten-targeting microRNAs. Additional PTEN loss in Pten,Pax5 double-mutant mice rescued FO B cell numbers and the development of MZ B cells but did not restore GC B cell formation. Hence, the posttranscriptional down-regulation of PTEN expression is an important function of Pax5 that facilitates the differentiation and survival of mature B cells, thereby promoting humoral immunity.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures



References
-
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:S45–S55. - PubMed
-
- Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011;11:34–46. - PubMed
-
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. - PubMed
-
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

