Spatiotemporal invasion dynamics of SARS-CoV-2 lineage B.1.1.7 emergence
- PMID: 34301854
- PMCID: PMC9269003
- DOI: 10.1126/science.abj0113
Spatiotemporal invasion dynamics of SARS-CoV-2 lineage B.1.1.7 emergence
Abstract
Understanding the causes and consequences of the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern is crucial to pandemic control yet difficult to achieve because they arise in the context of variable human behavior and immunity. We investigated the spatial invasion dynamics of lineage B.1.1.7 by jointly analyzing UK human mobility, virus genomes, and community-based polymerase chain reaction data. We identified a multistage spatial invasion process in which early B.1.1.7 growth rates were associated with mobility and asymmetric lineage export from a dominant source location, enhancing the effects of B.1.1.7's increased intrinsic transmissibility. We further explored how B.1.1.7 spread was shaped by nonpharmaceutical interventions and spatial variation in previous attack rates. Our findings show that careful accounting of the behavioral and epidemiological context within which variants of concern emerge is necessary to interpret correctly their observed relative growth rates.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



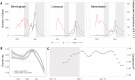
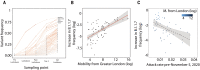
References
-
- M. Chand, S. Hopkins, C. Achison, C. Anderson, H. Allen, P. Blomquist, C. Chen, V. Chalker, G. Dabrera, O. Edeghere, M. Edmunds, T. Lamagni, R. Myers, I. Oliver, R. Elson, E. Gallagher, N. Groves, G. Hughes, M. Kall, H. Moore, W. Sopwith, C. Turner, L. Utsi, M. Vabistsevits, R. Vivancos, A. Zaidi, M. Zambon, W. Barclay, N. Ferguson, E. Volz, N. Loman, A. Rambaut, J. Barrett, Investigation of novel SARS-CoV-2 variant Variant of Concern 202012/01 (2021); https://assets.publishing.service.gov.uk/government/uploads/system/uploa....
-
- COVID-19 Genomics Consortium UK (CoG-UK), Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations (2020); https://virological.org/t/563.
-
- Alpert T., Brito A. F., Lasek-Nesselquist E., Rothman J., Valesano A. L., MacKay M. J., Petrone M. E., Breban M. I., Watkins A. E., Vogels C. B. F., Kalinich C. C., Dellicour S., Russell A., Kelly J. P., Shudt M., Plitnick J., Schneider E., Fitzsimmons W. J., Khullar G., Metti J., Dudley J. T., Nash M., Beaubier N., Wang J., Liu C., Hui P., Muyombwe A., Downing R., Razeq J., Bart S. M., Grills A., Morrison S. M., Murphy S., Neal C., Laszlo E., Rennert H., Cushing M., Westblade L., Velu P., Craney A., Cong L., Peaper D. R., Landry M. L., Cook P. W., Fauver J. R., Mason C. E., Lauring A. S., St George K., MacCannell D. R., Grubaugh N. D., Early introductions and transmission of SARS-CoV-2 variant B.1.1.7 in the United States. Cell 184, 2595–2604.e13 (2021). 10.1016/j.cell.2021.03.061 - DOI - PMC - PubMed
-
- O’Toole Á., Hill V., Pybus O. G., Watts A., Bogoch I. I., Khan K., Messina J. P., Tegally H., Lessells R. R., Giandhari J., Pillay S., Tumedi K. A., Nyepetsi G., Kebabonye M., Matsheka M., Mine M., Tokajian S., Hassan H., Salloum T., Merhi G., Koweyes J., Geoghegan J. L., de Ligt J., Ren X., Storey M., Freed N. E., Pattabiraman C., Prasad P., Desai A. S., Vasanthapuram R., Schulz T. F., Steinbrück L., Stadler T., Parisi A., Bianco A., García de Viedma D., Buenestado-Serrano S., Borges V., Isidro J., Duarte S., Gomes J. P., Zuckerman N. S., Mandelboim M., Mor O., Seemann T., Arnott A., Draper J., Gall M., Rawlinson W., Deveson I., Schlebusch S., McMahon J., Leong L., Lim C. K., Chironna M., Loconsole D., Bal A., Josset L., Holmes E., St George K., Lasek-Nesselquist E., Sikkema R. S., Oude Munnink B., Koopmans M., Brytting M., Sudha Rani V., Pavani S., Smura T., Heim A., Kurkela S., Umair M., Salman M., Bartolini B., Rueca M., Drosten C., Wolff T., Silander O., Eggink D., Reusken C., Vennema H., Park A., Carrington C., Sahadeo N., Carr M., Gonzalez G., de Oliveira T., Faria N., Rambaut A., Kraemer M. U. G., COVID-19 Genomics UK (COG-UK) consortium, Network for Genomic Surveillance in South Africa (NGS-SA), Brazil-UK CADDE Genomic Network, Swiss Viollier Sequencing Consortium, SEARCH Alliance San Diego, National Virus Reference Laboratory, SeqCOVID-Spain, Danish Covid-19 Genome Consortium (DCGC), Communicable Diseases Genomic Network (CDGN), Dutch National SARS-CoV-2 surveillance program, Division of Emerging Infectious Diseases (KDCA) , Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Wellcome Open Res. 6, 121 (2021). 10.12688/wellcomeopenres.16661.1 - DOI - PMC - PubMed
-
- Washington N. L., Gangavarapu K., Zeller M., Bolze A., Cirulli E. T., Schiabor Barrett K. M., Larsen B. B., Anderson C., White S., Cassens T., Jacobs S., Levan G., Nguyen J., Ramirez J. M. 3rd, Rivera-Garcia C., Sandoval E., Wang X., Wong D., Spencer E., Robles-Sikisaka R., Kurzban E., Hughes L. D., Deng X., Wang C., Servellita V., Valentine H., De Hoff P., Seaver P., Sathe S., Gietzen K., Sickler B., Antico J., Hoon K., Liu J., Harding A., Bakhtar O., Basler T., Austin B., MacCannell D., Isaksson M., Febbo P. G., Becker D., Laurent M., McDonald E., Yeo G. W., Knight R., Laurent L. C., de Feo E., Worobey M., Chiu C. Y., Suchard M. A., Lu J. T., Lee W., Andersen K. G., Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 184, 2587–2594.e7 (2021). 10.1016/j.cell.2021.03.052 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

