Pregnane X receptor exacerbates nonalcoholic fatty liver disease accompanied by obesity- and inflammation-prone gut microbiome signature
- PMID: 34303710
- PMCID: PMC9135326
- DOI: 10.1016/j.bcp.2021.114698
Pregnane X receptor exacerbates nonalcoholic fatty liver disease accompanied by obesity- and inflammation-prone gut microbiome signature
Abstract
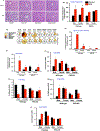
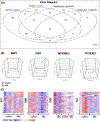
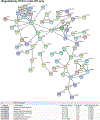
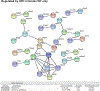
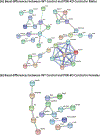
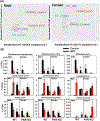
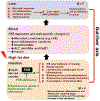
Nonalcoholic fatty liver disease (NAFLD) is the most prevalent chronic liver disease due to the current epidemics of obesity and diabetes. The pregnane X receptor (PXR) is a xenobiotic-sensing nuclear receptor known for trans-activating liver genes involved in drug metabolism and transport, and more recently implicated in energy metabolism. The gut microbiota can modulate the host xenobiotic biotransformation and contribute to the development of obesity. While the male sex confers a higher risk for NAFLD than women before menopause, the mechanism remains unknown. We hypothesized that the presence of PXR promotes obesity by modifying the gut-liver axis in a sex-specific manner. Male and female C57BL/6 (wild-type/WT) and PXR-knockout (PXR-KO) mice were fed control or high-fat diet (HFD) for 16-weeks. Serum parameters, liver histopathology, transcriptomic profiling, 16S-rDNA sequencing, and bile acid (BA) metabolomics were performed. PXR enhanced HFD-induced weight gain, hepatic steatosis and inflammation especially in males, accompanied by PXR-dependent up-regulation in hepatic genes involved in microbial response, inflammation, oxidative stress, and cancer; PXR-dependent increase in intestinal Firmicutes/Bacteroides ratio (hallmark of obesity) and the pro-inflammatory Lactobacillus, as well as a decrease in the anti-obese Allobaculum and the anti-inflammatory Bifidobacterum, with a PXR-dependent reduction of beneficial BAs in liver. The resistance to NAFLD in females may be explained by PXR-dependent decrease in pro-inflammatory bacteria (Ruminococcus gnavus and Peptococcaceae). In conclusion, PXR exacerbates hepatic steatosis and inflammation accompanied by obesity- and inflammation-prone gut microbiome signature, suggesting that gut microbiome may contribute to PXR-mediated exacerbation of NAFLD.
Keywords: Gut microbiome; High-fat diet; NAFLD; Obesity; PXR; Sex differences.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures









Similar articles
-
Pregnane X receptor knockout mitigates weight gain and hepatic metabolic dysregulation in female C57BL/6 J mice on a long-term high-fat diet.Biomed Pharmacother. 2024 Apr;173:116341. doi: 10.1016/j.biopha.2024.116341. Epub 2024 Feb 29. Biomed Pharmacother. 2024. PMID: 38428309 Free PMC article.
-
Protective effect of quercetin on high-fat diet-induced non-alcoholic fatty liver disease in mice is mediated by modulating intestinal microbiota imbalance and related gut-liver axis activation.Free Radic Biol Med. 2017 Jan;102:188-202. doi: 10.1016/j.freeradbiomed.2016.11.037. Epub 2016 Nov 25. Free Radic Biol Med. 2017. PMID: 27890642
-
Hepatocyte-specific SLC27A4 deletion ameliorates nonalcoholic fatty liver disease in mice via suppression of phosphatidylcholine-mediated PXR activation.Metabolism. 2025 Jan;162:156054. doi: 10.1016/j.metabol.2024.156054. Epub 2024 Nov 1. Metabolism. 2025. PMID: 39489412
-
Dual Role of Pregnane X Receptor in Nonalcoholic Fatty Liver Disease.Curr Mol Pharmacol. 2024;17:e18761429259143. doi: 10.2174/0118761429259143230927110556. Curr Mol Pharmacol. 2024. PMID: 37936450 Review.
-
Pregnane X Receptor and the Gut-Liver Axis: A Recent Update.Drug Metab Dispos. 2022 Apr;50(4):478-491. doi: 10.1124/dmd.121.000415. Epub 2021 Dec 3. Drug Metab Dispos. 2022. PMID: 34862253 Free PMC article. Review.
Cited by
-
Gut liver brain axis in diseases: the implications for therapeutic interventions.Signal Transduct Target Ther. 2023 Dec 6;8(1):443. doi: 10.1038/s41392-023-01673-4. Signal Transduct Target Ther. 2023. PMID: 38057297 Free PMC article. Review.
-
The Many Ages of Microbiome-Gut-Brain Axis.Nutrients. 2022 Jul 18;14(14):2937. doi: 10.3390/nu14142937. Nutrients. 2022. PMID: 35889894 Free PMC article.
-
Nidogen 2 Overexpression Promotes Hepatosteatosis and Atherosclerosis.Int J Mol Sci. 2024 Nov 28;25(23):12782. doi: 10.3390/ijms252312782. Int J Mol Sci. 2024. PMID: 39684493 Free PMC article.
-
Maternal PBDE exposure disrupts gut microbiome and promotes hepatic proinflammatory signaling in humanized PXR-transgenic mouse offspring over time.Toxicol Sci. 2023 Jul 28;194(2):209-225. doi: 10.1093/toxsci/kfad056. Toxicol Sci. 2023. PMID: 37267213 Free PMC article.
-
Research progress of gut microbiota and obesity caused by high-fat diet.Front Cell Infect Microbiol. 2023 Mar 13;13:1139800. doi: 10.3389/fcimb.2023.1139800. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36992691 Free PMC article. Review.
References
-
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M, Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes, Hepatology 64 (2016) 73–84. - PubMed
-
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI, An obesity-associated gut microbiome with increased capacity for energy harvest, Nature 444 (2006) 1027–1031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

