smFRET study of rRNA dimerization at the peptidyl transfer center
- PMID: 34303893
- PMCID: PMC8380723
- DOI: 10.1016/j.bpc.2021.106657
smFRET study of rRNA dimerization at the peptidyl transfer center
Abstract
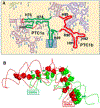
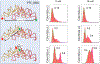
The ribosome is a ribozyme. At the peptidyl transfer center (PTC) of 180 nt, two loops (the A- and P- loops) bind to tRNAs and position them in close proximity for efficient peptidyl ligation. There is also a 2-fold rotational symmetry in the PTC, which suggests that the precursor of the modern ribosome possibly emerged through dimerization and gene fusion. However, experiments that demonstrate the possible dimerization have not yet been published. In our investigation, we reported single molecule FRET studies of two RNA fragments that generated high FRET values. By dye-labeling the 5'-biotinylated rRNA molecules at the 3'- terminals, or labeling three different types of tRNA-like oligos, we observed that RNA scaffolds can assemble and bring several short tRNA-acceptor-domain analogs, but not full-length tRNAs, to close proximity. Mg2+ and continuous 3-way junction motifs are essential to this process, but amino acid charging to the tRNA analogs is not required. We observed RNA dimers via native gel-shifting experiments. These experiments support the possible existence of a proto-ribosome in the form of an RNA dimer or multimer.
Keywords: Gel shifting; LUCA (last universal common ancestor); Peptide ligase; Peptidyl transferase activity; Single molecule FRET; tRNA-like oligo.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflicts of interest
There are no conflicts to declare.
Figures

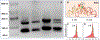


References
-
- Noller HF, Lancaster L, Mohan S, Zhou J, Ribosome structural dynamics in translocation: yet another functional role for ribosomal RNA, Q. Rev. Biophys, 50 (2017) e12. - PubMed
-
- Agmon I, Bashan A, Zarivach R, Yonath A, Symmetry at the active site of the ribosome: structural and functional implications, Biol Chem, 386 (2005) 833–844. - PubMed
-
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA, The structural basis of ribosome activity in peptide bond synthesis, Science, 289 (2000) 920–930. - PubMed
-
- Zhang B, Cech TR, Peptidyl-transferase ribozymes: trans reactions, structural characterization and ribosomal RNA-like features, Chem Biol, 5 (1998) 539–553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

