Breakers and amplifiers in chromatin circuitry: acetylation and ubiquitination control the heterochromatin machinery
- PMID: 34303934
- PMCID: PMC8667873
- DOI: 10.1016/j.sbi.2021.06.012
Breakers and amplifiers in chromatin circuitry: acetylation and ubiquitination control the heterochromatin machinery
Abstract
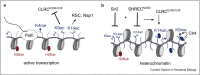
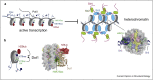
Eukaryotic genomes are segregated into active euchromatic and repressed heterochromatic compartments. Gene regulatory networks, chromosomal structures, and genome integrity rely on the timely and locus-specific establishment of active and silent states to protect the genome and provide the basis for cell division and specification of cellular identity. Here, we focus on the mechanisms and molecular machinery that establish heterochromatin in Schizosaccharomyces pombe and compare it with Saccharomyces cerevisiae and the mammalian polycomb system. We present recent structural and mechanistic evidence, which suggests that histone acetylation protects active transcription by disrupting the positive feedback loops used by the heterochromatin machinery and that H2A and H3 monoubiquitination actively drives heterochromatin, whereas H2B monoubiquitination mobilizes the defenses to quench heterochromatin.
Copyright © 2021 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Conflict of interest statement Nothing declared.
Figures


Similar articles
-
The C-terminus of histone H2B is involved in chromatin compaction specifically at telomeres, independently of its monoubiquitylation at lysine 123.PLoS One. 2011;6(7):e22209. doi: 10.1371/journal.pone.0022209. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829450 Free PMC article.
-
The interplay of histone H2B ubiquitination with budding and fission yeast heterochromatin.Curr Genet. 2018 Aug;64(4):799-806. doi: 10.1007/s00294-018-0812-1. Epub 2018 Feb 20. Curr Genet. 2018. PMID: 29464330 Free PMC article. Review.
-
Recruitment and allosteric stimulation of a histone-deubiquitinating enzyme during heterochromatin assembly.J Biol Chem. 2018 Feb 16;293(7):2498-2509. doi: 10.1074/jbc.RA117.000498. Epub 2017 Dec 29. J Biol Chem. 2018. PMID: 29288197 Free PMC article.
-
The Histone Acetyltransferase Mst2 Protects Active Chromatin from Epigenetic Silencing by Acetylating the Ubiquitin Ligase Brl1.Mol Cell. 2017 Jul 20;67(2):294-307.e9. doi: 10.1016/j.molcel.2017.05.026. Epub 2017 Jun 22. Mol Cell. 2017. PMID: 28648780 Free PMC article.
-
Epigenetics in Saccharomyces cerevisiae.Cold Spring Harb Perspect Biol. 2013 Jul 1;5(7):a017491. doi: 10.1101/cshperspect.a017491. Cold Spring Harb Perspect Biol. 2013. PMID: 23818500 Free PMC article. Review.
Cited by
-
Context dependent function of the transcriptional regulator Rap1 in gene silencing and activation in Saccharomyces cerevisiae.bioRxiv [Preprint]. 2023 May 11:2023.05.08.539937. doi: 10.1101/2023.05.08.539937. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Oct 3;120(40):e2304343120. doi: 10.1073/pnas.2304343120. PMID: 37214837 Free PMC article. Updated. Preprint.
-
Context-dependent function of the transcriptional regulator Rap1 in gene silencing and activation in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2023 Oct 3;120(40):e2304343120. doi: 10.1073/pnas.2304343120. Epub 2023 Sep 28. Proc Natl Acad Sci U S A. 2023. PMID: 37769255 Free PMC article.
-
Increased DNA damage in full-grown oocytes is correlated with diminished autophagy activation.Nat Commun. 2024 Nov 1;15(1):9463. doi: 10.1038/s41467-024-53559-w. Nat Commun. 2024. PMID: 39487138 Free PMC article.
-
Bordetella adenylate cyclase toxin elicits chromatin remodeling and transcriptional reprogramming that blocks differentiation of monocytes into macrophages.mBio. 2025 Apr 9;16(4):e0013825. doi: 10.1128/mbio.00138-25. Epub 2025 Mar 19. mBio. 2025. PMID: 40105369 Free PMC article.
-
Oncogenic IDH1mut drives robust loss of histone acetylation and increases chromatin heterogeneity.Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2403862122. doi: 10.1073/pnas.2403862122. Epub 2024 Dec 30. Proc Natl Acad Sci U S A. 2025. PMID: 39793065 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

