Making waves: Defining the lead time of wastewater-based epidemiology for COVID-19
- PMID: 34304074
- PMCID: PMC8282235
- DOI: 10.1016/j.watres.2021.117433
Making waves: Defining the lead time of wastewater-based epidemiology for COVID-19
Abstract
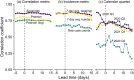
Individuals infected with SARS-CoV-2, the virus that causes COVID-19, may shed the virus in stool before developing symptoms, suggesting that measurements of SARS-CoV-2 concentrations in wastewater could be a "leading indicator" of COVID-19 prevalence. Multiple studies have corroborated the leading indicator concept by showing that the correlation between wastewater measurements and COVID-19 case counts is maximized when case counts are lagged. However, the meaning of "leading indicator" will depend on the specific application of wastewater-based epidemiology, and the correlation analysis is not relevant for all applications. In fact, the quantification of a leading indicator will depend on epidemiological, biological, and health systems factors. Thus, there is no single "lead time" for wastewater-based COVID-19 monitoring. To illustrate this complexity, we enumerate three different applications of wastewater-based epidemiology for COVID-19: a qualitative "early warning" system; an independent, quantitative estimate of disease prevalence; and a quantitative alert of bursts of disease incidence. The leading indicator concept has different definitions and utility in each application.
Keywords: COVID-19; SARS-CoV-2; Surveillance; Wastewater-based epidemiology.
Copyright © 2021. Published by Elsevier Ltd.
Conflict of interest statement
None.
Figures
References
-
- Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 Aug 1138764doi: 10.1016/j.scitotenv.2020.138764. Epub 2020 Apr 18. PMID: 32387778; PMCID: PMC7165106. - PMC - PubMed
-
- Ahmed W, Bertsch PM, Angel N, Bibby K, Bivins A, Dierens L, Edson J, Ehret J, Gyawali P, Hamilton KA, Hosegood I, Hugenholtz P, Jiang G, Kitajima M, Sichani HT, Shi J, Shimko KM, Simpson SL, Smith WJM, Symonds EM, Thomas KV, Verhagen R, Zaugg J, Mueller JF. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020;27(5) Aug 20taaa116. doi: 10.1093/jtm/taaa116. PMID: 32662867; PMCID: PMC7454825. - PMC - PubMed
-
- Betancourt WW, Schmitz BW, Innes GK, Pogreba Brown KM, Prasek SM, Stark ER, Foster AR, Sprissler RS, Harris DT, Sherchan SP, Gerba CP, Pepper IL. Cold Spring Harbor Laboratory; 2020. Wastewater-based Epidemiology for Averting COVID-19 Outbreaks on The University of Arizona Campus. Nov 16Available from: http://dx.doi.org/ - DOI
-
- Bloom RM, Buckeridge DL, Cheng KE. Finding Leading Indicators for Disease Outbreaks: Filtering, Cross-correlation, and Caveats. Journal of the American Medical Informatics Association. 2007;14(1):76–85. doi: 10.1197/jamia.m2178. [Internet]. Oxford University Press (OUP)Jan 1Available from: http://dx.doi.org/ - DOI - PMC - PubMed
-
- Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. Vol. 2. Elsevier BV; 2021. pp. e13–e22. (SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. The Lancet Microbe [Internet]). JanAvailable fromhttp://dx.doi.org/ - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

