Targeting SARS-CoV-2 main protease by teicoplanin: A mechanistic insight by docking, MM/GBSA and molecular dynamics simulation
- PMID: 34305175
- PMCID: PMC8286173
- DOI: 10.1016/j.molstruc.2021.131124
Targeting SARS-CoV-2 main protease by teicoplanin: A mechanistic insight by docking, MM/GBSA and molecular dynamics simulation
Abstract
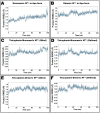
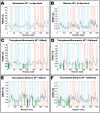
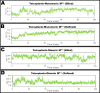
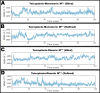
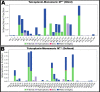
First emerged in late December 2019, the outbreak of novel severe acute respiratory syndrome corona virus-2 (SARS-CoV-2) pandemic has instigated public-health emergency around the globe. Till date there is no specific therapeutic agent for this disease and hence, the world is craving to identify potential antiviral agents against SARS-CoV-2. The main protease (MPro) is considered as an attractive drug target for rational drug design against SARS-CoV-2 as it is known to play a crucial role in the viral replication and transcription. Teicoplanin is a glycopeptide class of antibiotic which is regularly used for treating Gram-positive bacterial infections, has shown potential therapeutic efficacy against SARS-CoV-2 in vitro. Therefore, in this study, a mechanistic insight of intermolecular interactions between teicoplanin and SARS-CoV-2 MPro has been scrutinized by molecular docking. Both monomeric and dimeric forms of MPro was used in docking involving blind as well as defined binding site based on the known inhibitor. Binding energies of teicoplanin-MPro complexes were estimated by Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) computations from docking and simulated trajectories. The dynamic and thermodynamics constraints of docked drug in complex with target proteins under specific physiological conditions was ascertained by all-atom molecular dynamics simulation of 100 ns trajectory. Root mean square deviation and fluctuation of carbon α chain justified the stability of the bound complex in biological environments. The outcomes of current study are supposed to be fruitful in rational design of antiviral drugs against SARS-CoV-2.
Keywords: Covid-19; Docking; Molecular dynamics; SARS-CoV-2 main protease; Teicoplanin.
© 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Hui D.S., Azhar E.I, Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. - DOI - PMC - PubMed
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
-
- Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., Haque U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 2020;49:717–726. doi: 10.1093/ije/dyaa033. - DOI - PMC - PubMed
-
- Tsai P.-H., Lai W.-Y., Lin Y.-Y., Luo Y.-H., Lin Y.-T., Chen H.-K., Chen Y.-M., Lai Y.-C., Kuo L.-C., Chen S.-D., Chang K.-J., Liu C.-H., Chang S.-C., Wang F.-D., Yang Y.-P. Clinical manifestation and disease progression in COVID-19 infection. J. Chin. Med. Assoc. 2021;84 https://journals.lww.com/jcma/Fulltext/2021/01000/Clinical_manifestation... - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

