Global Transcriptomics Uncovers Distinct Contributions From Splicing Regulatory Proteins to the Macrophage Innate Immune Response
- PMID: 34305890
- PMCID: PMC8299563
- DOI: 10.3389/fimmu.2021.656885
Global Transcriptomics Uncovers Distinct Contributions From Splicing Regulatory Proteins to the Macrophage Innate Immune Response
Abstract
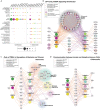
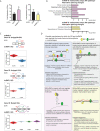
Pathogen sensing via pattern recognition receptors triggers massive reprogramming of macrophage gene expression. While the signaling cascades and transcription factors that activate these responses are well-known, the role of post-transcriptional RNA processing in modulating innate immune gene expression remains understudied. Given their crucial role in regulating pre-mRNA splicing and other RNA processing steps, we hypothesized that members of the SR/hnRNP protein families regulate innate immune gene expression in distinct ways. We analyzed steady state gene expression and alternatively spliced isoform production in ten SR/hnRNP knockdown RAW 264.7 macrophage-like cell lines following infection with the bacterial pathogen Salmonella enterica serovar Typhimurium (Salmonella). We identified thousands of transcripts whose abundance is increased or decreased by SR/hnRNP knockdown in macrophages. Notably, we observed that SR and hnRNP proteins influence expression of different genes in uninfected versus Salmonella-infected macrophages, suggesting functionalization of these proteins upon pathogen sensing. Likewise, we found that knockdown of SR/hnRNPs promoted differential isoform usage (DIU) for thousands of macrophage transcripts and that these alternative splicing changes were distinct in uninfected and Salmonella-infected macrophages. Finally, having observed a surprising degree of similarity between the differentially expressed genes (DEGs) and DIUs in hnRNP K and U knockdown macrophages, we found that hnRNP K and U knockdown macrophages are both more restrictive to Vesicular Stomatitis Virus (VSV), while hnRNP K knockdown macrophages are more permissive to Salmonella Typhimurium. Based on these findings, we conclude that many innate immune genes evolved to rely on one or more SR/hnRNPs to ensure the proper magnitude of their induction, supporting a model wherein pre-mRNA splicing is critical for regulating innate immune gene expression and controlling infection outcomes in macrophages ex vivo.
Keywords: RNA binding protein; SR protein; Salmonella Typhimurium; hnRNP; inflammation; pre-mRNA splicing.
Copyright © 2021 Wagner, Scott, West, Vail, Fitzsimons, Coleman, Carter, Watson and Patrick.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
The Splicing Factor hnRNP M Is a Critical Regulator of Innate Immune Gene Expression in Macrophages.Cell Rep. 2019 Nov 5;29(6):1594-1609.e5. doi: 10.1016/j.celrep.2019.09.078. Cell Rep. 2019. PMID: 31693898 Free PMC article.
-
Toll-Like receptor 2 (TLR2) and TLR9 play opposing roles in host innate immunity against Salmonella enterica serovar Typhimurium infection.Infect Immun. 2015 Apr;83(4):1641-9. doi: 10.1128/IAI.02870-14. Epub 2015 Feb 9. Infect Immun. 2015. PMID: 25667264 Free PMC article.
-
Neutrophil prime unique transcriptional responses in intestinal organoids during infection with nontyphoidal Salmonella enterica serovars.mSphere. 2024 Dec 19;9(12):e0069324. doi: 10.1128/msphere.00693-24. Epub 2024 Nov 20. mSphere. 2024. PMID: 39565098 Free PMC article.
-
Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer.Cancer Lett. 2017 Jun 28;396:53-65. doi: 10.1016/j.canlet.2017.03.013. Epub 2017 Mar 14. Cancer Lett. 2017. PMID: 28315432 Review.
-
Critical role of type I interferon-induced macrophage necroptosis during infection with Salmonella enterica serovar Typhimurium.Cell Mol Immunol. 2013 Mar;10(2):99-100. doi: 10.1038/cmi.2012.68. Epub 2012 Dec 24. Cell Mol Immunol. 2013. PMID: 23262973 Free PMC article. Review. No abstract available.
Cited by
-
ERH regulates type II interferon immune signaling through post-transcriptional regulation of JAK2 mRNA.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf545. doi: 10.1093/nar/gkaf545. Nucleic Acids Res. 2025. PMID: 40586312 Free PMC article.
-
A Bioinformatics Approach to Identifying Potential Biomarkers for Cryptosporidium parvum: A Coccidian Parasite Associated with Fetal Diarrhea.Vaccines (Basel). 2021 Dec 2;9(12):1427. doi: 10.3390/vaccines9121427. Vaccines (Basel). 2021. PMID: 34960172 Free PMC article.
-
RNA regulatory mechanisms that control antiviral innate immunity.Immunol Rev. 2021 Nov;304(1):77-96. doi: 10.1111/imr.13019. Epub 2021 Aug 17. Immunol Rev. 2021. PMID: 34405416 Free PMC article. Review.
-
Adipocyte-specific Steap4 deficiency reduced thermogenesis and energy expenditure in mice.iScience. 2025 Jan 25;28(2):111903. doi: 10.1016/j.isci.2025.111903. eCollection 2025 Feb 21. iScience. 2025. PMID: 39995871 Free PMC article.
-
Serine/arginine-rich splicing factor 7 promotes the type I interferon response by activating Irf7 transcription.Cell Rep. 2024 Mar 26;43(3):113816. doi: 10.1016/j.celrep.2024.113816. Epub 2024 Feb 22. Cell Rep. 2024. PMID: 38393946 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

