Molecular Insights of Nickel Binding to Therapeutic Antibodies as a Possible New Antibody Superantigen
- PMID: 34305906
- PMCID: PMC8296638
- DOI: 10.3389/fimmu.2021.676048
Molecular Insights of Nickel Binding to Therapeutic Antibodies as a Possible New Antibody Superantigen
Abstract
The binding of nickel by immune proteins can manifest as Type IV contact dermatitis (Ni-specific T cells mediated) and less frequently as Type I hypersensitivity with both mechanisms remaining unknown to date. Since there are reports of patients co-manifesting the two hypersensitivities, a common mechanism may underlie both the TCR and IgE nickel binding. Focusing on Trastuzumab and Pertuzumab IgE variants as serendipitous investigation models, we found Ni-NTA interactions independent of Her2 binding to be due to glutamine stretches. These stretches are both Ni-inducible and in fixed pockets at the antibody complementarity-determining regions (CDRs) and framework regions (FWRs) of both the antibody heavy and light chains with influence from the heavy chain constant region. Comparisons with TCRs structures revealed similar interactions, demonstrating the possible underlying mechanism in selecting for Ni-binding IgEs and TCRs respectively. With the elucidation of the interaction, future therapeutic antibodies could also be sagaciously engineered to utilize such nickel binding for biotechnological purposes.
Keywords: IgE; IgG1; allergy; antibody; glutamine; nickel (II); type I hypersensitivity.
Copyright © 2021 Su, Lua, Poh, Ling, Yeo and Gan.
Conflict of interest statement
SKEG was employed by APD SKEG Pte Ltd., Singapore, Singapore. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

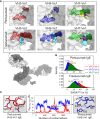


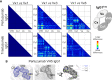
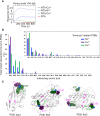
References
-
- Gelardi M, Guarino R, Taliente S, Quaranta N, Carperntieri A, Passalacqua G. Allergic and Nonallergic Rhinitis and Skin Sensitization to Metals: Is There a Link? Eur Ann Allergy Clin Immunol (2017) 49(3):106–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

