SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns
- PMID: 34305949
- PMCID: PMC8293742
- DOI: 10.3389/fimmu.2021.708264
SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns
Abstract
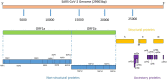
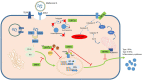
There are still many unanswered questions concerning viral SARS-CoV-2 pathogenesis in COVID-19. Accessory proteins in SARS-CoV-2 consist of eleven viral proteins whose roles during infection are still not completely understood. Here, a review on the current knowledge of SARS-CoV-2 accessory proteins is summarized updating new research that could be critical in understanding SARS-CoV-2 interaction with the host. Some accessory proteins such as ORF3b, ORF6, ORF7a and ORF8 have been shown to be important IFN-I antagonists inducing an impairment in the host immune response. In addition, ORF3a is involved in apoptosis whereas others like ORF9b and ORF9c interact with cellular organelles leading to suppression of the antiviral response in infected cells. However, possible roles of ORF7b and ORF10 are still awaiting to be described. Also, ORF3d has been reassigned. Relevant information on the knowns and the unknowns in these proteins is analyzed, which could be crucial for further understanding of SARS-CoV-2 pathogenesis and to design strategies counteracting their actions evading immune responses in COVID-19.
Keywords: COVID-19; SARS-CoV-2; accessory proteins; coronavirus; immune response.
Copyright © 2021 Redondo, Zaldívar-López, Garrido and Montoya.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous