Microbial Identification, High-Resolution Microscopy and Spectrometry of the Rhizosphere in Its Native Spatial Context
- PMID: 34305970
- PMCID: PMC8293618
- DOI: 10.3389/fpls.2021.668929
Microbial Identification, High-Resolution Microscopy and Spectrometry of the Rhizosphere in Its Native Spatial Context
Erratum in
-
Corrigendum: Microbial identification, high-resolution microscopy and spectrometry of the rhizosphere in its native spatial context.Front Plant Sci. 2023 Jan 9;13:1125001. doi: 10.3389/fpls.2022.1125001. eCollection 2022. Front Plant Sci. 2023. PMID: 36699837 Free PMC article.
Abstract
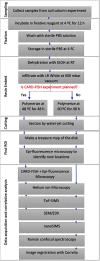
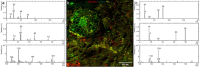
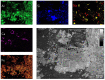
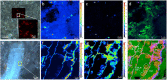
During the past decades, several stand-alone and combinatorial methods have been developed to investigate the chemistry (i.e., mapping of elemental, isotopic, and molecular composition) and the role of microbes in soil and rhizosphere. However, none of these approaches are currently applicable to characterize soil-root-microbe interactions simultaneously in their spatial arrangement. Here we present a novel approach that allows for simultaneous microbial identification and chemical analysis of the rhizosphere at micro- to nano-meter spatial resolution. Our approach includes (i) a resin embedding and sectioning method suitable for simultaneous correlative characterization of Zea mays rhizosphere, (ii) an analytical work flow that allows up to six instruments/techniques to be used correlatively, and (iii) data and image correlation. Hydrophilic, immunohistochemistry compatible, low viscosity LR white resin was used to embed the rhizosphere sample. We employed waterjet cutting and avoided polishing the surface to prevent smearing of the sample surface at nanoscale. The quality of embedding was analyzed by Helium Ion Microscopy (HIM). Bacteria in the embedded soil were identified by Catalyzed Reporter Deposition-Fluorescence in situ Hybridization (CARD-FISH) to avoid interferences from high levels of autofluorescence emitted by soil particles and organic matter. Chemical mapping of the rhizosphere was done by Scanning Electron Microscopy (SEM) with Energy-dispersive X-ray analysis (SEM-EDX), Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS), nano-focused Secondary Ion mass Spectrometry (nanoSIMS), and confocal Raman spectroscopy (μ-Raman). High-resolution correlative characterization by six different techniques followed by image registration shows that this method can meet the demanding requirements of multiple characterization techniques to identify spatial organization of bacteria and chemically map the rhizosphere. Finally, we presented individual and correlative workflows for imaging and image registration to analyze data. We hope this method will be a platform to combine various 2D analytics for an improved understanding of the rhizosphere processes and their ecological significance.
Keywords: CARD-FISH; Helium Ion Microscopy; London Resin White embedding; Rhizosphere; Secondary Ion Mass Spectrometry; Soil bacteria; Water-jet cutting; correlative chemical microscopy.
Copyright © 2021 Bandara, Schmidt, Davoudpour, Stryhanyuk, Richnow and Musat.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Bandara C. D., Schmidt M., Davoudpour Y., Stryhanyuk H., Richnow H. H., Musat N. (2021). High-resolution chemical mapping and microbial identification of rhizosphere using correlative microscopy. bioRxiv 2021:429689. - PubMed
-
- Baveye P. C., Otten W., Kravchenko A., Balseiro-Romero M., Beckers E., Chalhoub M., et al. (2018). Emergent properties of microbial activity in heterogeneous soil microenvironments: different research approaches are slowly converging, yet major challenges remain. Front. Microbiol. 9:1929. 10.3389/fmicb.2018.01929 - DOI - PMC - PubMed
-
- Benettoni P., Stryhanyuk H., Wagner S., Kollmer F., Moreno Osorio J. H., Schmidt M., et al. (2019). Identification of nanoparticles and their localization in algal biofilm by 3D-imaging secondary ion mass spectrometry. J. Analyt. Atomic Spectrometry 34 1098–1108. 10.1039/c8ja00439k - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

