Analysis of immune subtypes across the epithelial-mesenchymal plasticity spectrum
- PMID: 34306571
- PMCID: PMC8283019
- DOI: 10.1016/j.csbj.2021.06.023
Analysis of immune subtypes across the epithelial-mesenchymal plasticity spectrum
Abstract
Epithelial-mesenchymal plasticity plays a critical role in many solid tumor types as a mediator of metastatic dissemination and treatment resistance. In addition, there is also a growing appreciation that the epithelial/mesenchymal status of a tumor plays a role in immune evasion and immune suppression. A deeper understanding of the immunological features of different tumor types has been facilitated by the availability of large gene expression datasets and the development of methods to deconvolute bulk RNA-Seq data. These resources have generated powerful new ways of characterizing tumors, including classification of immune subtypes based on differential expression of immunological genes. In the present work, we combine scoring algorithms to quantify epithelial-mesenchymal plasticity with immune subtype analysis to understand the relationship between epithelial plasticity and immune subtype across cancers. We find heterogeneity of epithelial-mesenchymal transition (EMT) status both within and between cancer types, with greater heterogeneity in the expression of EMT-related factors than of MET-related factors. We also find that specific immune subtypes have associated EMT scores and differential expression of immune checkpoint markers.
Keywords: Gene expression signature; Hybrid epithelial/mesenchymal cells; Immune invasion; Immune subtypes; Tumor heterogeneity.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
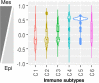

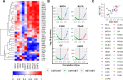

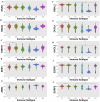

References
-
- Nieto M.A., Huang R.-Y.-J., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166(1):21–45. - PubMed
-
- Yang, J., Antin, P., Berx, G., Blanpain, C., Brabletz, T., Bronner, M., Campbell, K., Cano, A., Casanova, J., Christofori, G., Dedhar, S., Derynck, R., Ford, H. L., Fuxe, J., García de Herreros, A., Goodall, G. J., Hadjantonakis, A.-K., Huang, R. J. Y., Kalcheim, C., … EMT International Association (TEMTIA). (2020). Guidelines and definitions for research on epithelial-mesenchymal transition. Nature Reviews. Molecular Cell Biology, 21(6), 341–352. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
