Pan-Cancer Survey of Tumor Mass Dormancy and Underlying Mutational Processes
- PMID: 34307377
- PMCID: PMC8299471
- DOI: 10.3389/fcell.2021.698659
Pan-Cancer Survey of Tumor Mass Dormancy and Underlying Mutational Processes
Abstract
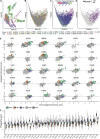
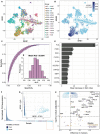
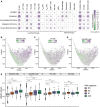
Tumor mass dormancy is the key intermediate step between immune surveillance and cancer progression, yet due to its transitory nature it has been difficult to capture and characterize. Little is understood of its prevalence across cancer types and of the mutational background that may favor such a state. While this balance is finely tuned internally by the equilibrium between cell proliferation and cell death, the main external factors contributing to tumor mass dormancy are immunological and angiogenic. To understand the genomic and cellular context in which tumor mass dormancy may develop, we comprehensively profiled signals of immune and angiogenic dormancy in 9,631 cancers from the Cancer Genome Atlas and linked them to tumor mutagenesis. We find evidence for immunological and angiogenic dormancy-like signals in 16.5% of bulk sequenced tumors, with a frequency of up to 33% in certain tissues. Mutations in the CASP8 and HRAS oncogenes were positively selected in dormant tumors, suggesting an evolutionary pressure for controlling cell growth/apoptosis signals. By surveying the mutational damage patterns left in the genome by known cancer risk factors, we found that aging-induced mutations were relatively depleted in these tumors, while patterns of smoking and defective base excision repair were linked with increased tumor mass dormancy. Furthermore, we identified a link between APOBEC mutagenesis and dormancy, which comes in conjunction with immune exhaustion and may partly depend on the expression of the angiogenesis regulator PLG as well as interferon and chemokine signals. Tumor mass dormancy also appeared to be impaired in hypoxic conditions in the majority of cancers. The microenvironment of dormant cancers was enriched in cytotoxic and regulatory T cells, as expected, but also in macrophages and showed a reduction in inflammatory Th17 signals. Finally, tumor mass dormancy was linked with improved patient survival outcomes. Our analysis sheds light onto the complex interplay between dormancy, exhaustion, APOBEC activity and hypoxia, and sets directions for future mechanistic explorations.
Keywords: APOBEC; angiogenesis; hypoxia; immunity; mutational signatures; tumor mass dormancy.
Copyright © 2021 Wiecek, Jacobson, Lason and Secrier.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Aguirre-Ghiso J. A., Estrada Y., Liu D., Ossowski L. (2003). ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK). Cancer Res. 63 1684–1695. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

