Epigenetic Mechanisms in DNA Double Strand Break Repair: A Clinical Review
- PMID: 34307454
- PMCID: PMC8292790
- DOI: 10.3389/fmolb.2021.685440
Epigenetic Mechanisms in DNA Double Strand Break Repair: A Clinical Review
Abstract
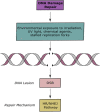
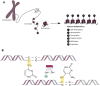
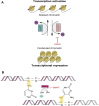
Upon the induction of DNA damage, the chromatin structure unwinds to allow access to enzymes to catalyse the repair. The regulation of the winding and unwinding of chromatin occurs via epigenetic modifications, which can alter gene expression without changing the DNA sequence. Epigenetic mechanisms such as histone acetylation and DNA methylation are known to be reversible and have been indicated to play different roles in the repair of DNA. More importantly, the inhibition of such mechanisms has been reported to play a role in the repair of double strand breaks, the most detrimental type of DNA damage. This occurs by manipulating the chromatin structure and the expression of essential proteins that are critical for homologous recombination and non-homologous end joining repair pathways. Inhibitors of histone deacetylases and DNA methyltransferases have demonstrated efficacy in the clinic and represent a promising approach for cancer therapy. The aims of this review are to summarise the role of histone deacetylase and DNA methyltransferase inhibitors involved in DNA double strand break repair and explore their current and future independent use in combination with other DNA repair inhibitors or pre-existing therapies in the clinic.
Keywords: DNA double strand breaks; DNA methyltransferase inhibitors; DNA repair; epigenetic mechanisms; histone deacetylase inhibitors.
Copyright © 2021 Fernandez, O’Leary, O’Byrne, Burgess, Richard and Suraweera.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abbotts R., Topper M. J., Biondi C., Fontaine D., Goswami R., Stojanovic L., et al. (2019). DNA Methyltransferase Inhibitors Induce a BRCAness Phenotype that Sensitizes NSCLC to PARP Inhibitor and Ionizing Radiation. Proc. Natl. Acad. Sci. USA 116 (45), 22609–22618. 10.1073/pnas.1903765116 - DOI - PMC - PubMed
-
- Al-Hamamah M. A., Alotaibi M. R., Ahmad S. F., Ansari M. A., Attia M. S. M., Nadeem A., et al. (2019). Genetic and Epigenetic Alterations Induced by the Small-Molecule Panobinostat: A Mechanistic Study at the Chromosome and Gene Levels. DNA Repair 78 (June), 70–80. 10.1016/j.dnarep.2019.03.008, - DOI - PubMed
-
- Alexandrova E., Lamberti J., Saggese P., Pecoraro G., Memoli D., Valeria Mirici C., et al. (2020). Small Non-coding RNA Profiling Identifies MiR-181a-5p as a Mediator of Estrogen Receptor Beta-Induced Inhibition of Cholesterol Biosynthesis in Triple-Negative Breast Cancer. Cells 9 (4), 874. 10.3390/cells9040874 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

